Abstract
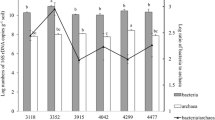
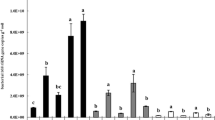
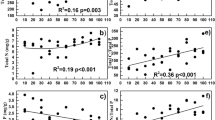
The integrated biomass beneath the surface horizon in unsaturated soils is large and potentially important in nutrient and carbon cycling. Compared to surface soils, the ecology of these subsurface soils is weakly understood, particularly in terms of the composition of bacterial communities. We compared soil bacterial communities along two vertical transects by terminal restriction fragment length polymorphisms (TRFLPs) of PCR-amplified 16S rRNA genes to determine how surface and deep bacterial communities differ. DNA yield from soils collected from two Mediterranean grassland transects decreased exponentially from the surface to 4 m deep. Richness, as assessed by the number of peaks obtained after restriction with HhaI, MspI, RsaI, or HaeIII, and diversity, as assessed by the Shannon diversity indices, were lowest in the deepest sample. Lower diversity at depth is consistent with species-energy theory, which would predict relatively low diversity in the low organic matter horizons. Principal components analysis suggested that, in terms of HhaI and HaeIII generated TRFLPs, bacterial communities differed between depths. The most abundant amplicons cloned from the deepest sample contained sequences with restriction sites consistent with the largest peaks observed in TRFLPs generated from deep samples. These more abundant operational taxonomic units (OTUs) appeared related to Pseudomonas and Variovorax. Several OTUs were more related to each other than any previously described ribotypes. These OTUs showed similarity to bacteria from the divisions Actinobacteria and Firmicutes.
Similar content being viewed by others
References
Ajwa HA, Rice CW, Sotomayor D (1998) Carbon and nitrogen mineralization in tallgrass prairie and agricultural soil profiles. Soil Sci Soc Am J 62:942–951
Atlas RM, Bartha R (1993) Microbial Ecology: Fundamentals and Applications, 3rd ed. Benjamin/Cummings, Redwood City, CA
Balkwill DL, Reeves RH, Drake GR, Reeves JY, Crocker FH, King MB, Boone DR (1997) Phylogenetic characterization of bacteria in the subsurface microbial culture collection. FEMS Microbiol Rev 20:201–216
Barns SM, Takala SL, Kuske CR (1999) Wide distribution and diversity of members of the bacterial kingdom Acidobacterium in the environment. Appl Environ Microbiol 65:1731–1737
Batjes NH (1996) Total carbon and nitrogen in soils of the world. Eur J Soil Sci 47:151–163
Blume E, Bischoff M, Reichert JM, Moorman T, Konopka A, Turco RF (2002) Surface and subsurface microbial biomass, community structure and metabolic activity as a function of soil depth and season. Appl Soil Ecol 20:171–181
Brockman FJ, Li SW, Fredrickson JK, Ringelberg DB, Kieft TL, Spadoni CM, White DC, McKinley JP (1998) Post-sampling changes in microbial community composition and activity in a subsurface paleosol. Microb Ecol 36:152–164
Brodie E, Edwards S, Clipson N (2002) Bacterial community dynamics across a floristic gradient in a temperate upland grassland ecosystem. Microb Ecol 44:260–270
Buckley DH, Schmidt TM (2001) The structure of microbial communities in soil and the lasting impact of cultivation. Microb Ecol 42:11–21
Chandler DP, Brockman FJ, Bailey TJ, Fredrickson JK (1998) Phylogenetic diversity of archaea and bacteria in a deep subsurface paleosol. Microb Ecol 36:37–50
Chandler DP, Brockman FJ, Fredrickson JK (1997) Use of 16S rDNA clone libraries to study changes in a microbial community resulting from ex situ perturbation of a subsurface sediment. FEMS Microbiol Rev 20:217–230
Crocker FH, Fredrickson JK, White DC, Ringelberg DB, Balkwill DL (2000) Phylogenetic and physiological diversity of Arthrobacter strains isolated from unconsolidated subsurface sediments. Microbiology-(UK) 146:1295–1310
Curtis TP, Sloan WT, Scannell JW (2002) Estimating prokaryotic diversity and its limits. Proc Natl Acad Sci USA 99:10494–10499
Dunbar J, Ticknor LO, Kuske CR (2000) Assessment of microbial diversity in four southwestern United States soils by 16S rRNA gene terminal restriction fragment analysis. Appl Environ Microbiol 66:2943–2950
Dunbar J, Ticknor LO, Kuske CR (2001) Phylogenetic specificity and reproducibility and new method for analysis of terminal restriction fragment profiles of 16S rRNA genes from bacterial communities. Appl Environ Microbiol 67:190–197
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Felsenstein J (1989) PHYLIP-Phylogeny Inference Package (Version 3.2). Cladistics 5:164–166
Fierer N, Schimel JP, Holden PA (2003) Influence of drying-rewetting frequency on soil bacterial community structure. Microb Ecol 45:63–71
Fierer N, Schimel JP, Holden PA (2003) Variations in microbial community composition through two soil depth profiles. Soil Biol Biochem 35:167–176
Fredrickson JK, Brockman FJ, Bjornstad BN, Long PE, Li SW, McKinley JP, Wright JV, Conca JL, Kieft TL, Balkwill DL (1993) Microbiological characteristics of pristine and contaminated deep vadose sediments from an arid region. Geomicrobiol J 11:95–107
Frostegärd Å, Courtois S, Ramisse V, Clerc S, Bernillon D, Le Gall F, Jeannin P, Nesme X, Simonet P (1999) Quantification of bias related to the extraction of DNA directly from soils. Appl Environ Microbiol 65:5409–5420
Hamilton JG, Holzapfel C, Mahall BE (1999) Coexistence and interference between a native perennial grass and non-native annual grasses in California. Oecologia 121:518–526
Head IM, Saunders JR, Pickup RW (1998) Microbial evolution, diversity, and ecology: A decade of ribosomal RNA analysis of uncultivated microorganisms. Microb Ecol 35:1–21
Hedrick DB, Peacock A, Stephen JR, Macnaughton SJ, Brüggemann J, White DC (2000) Measuring soil microbial community diversity using polar lipid fatty acid and denaturing gradient gel electrophoresis data. J Microbiol Methods 41:235–248
Holben WE, Jansson JK, Chelm BK, Tiedje JM (1988) DNA probe method for the detection of specific microorganisms in the soil bacterial community. Appl Environ Microbiol 54:703–711
Hugenholtz P, Goebel BM, Pace NR (1998) Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol 180:4765–4774
Jackson CR, Harper JP, Willoughby D, Roden EE, Churchill PF (1997) A simple, efficient method for the separation of humic substances and DNA from environmental samples. Appl Environ Microbiol 63:4993–4995
Kieft TL, Wilch E, O’connor K, Ringelberg DB, White DC (1997) Survival and phospholipid fatty acid profiles of surface and subsurface bacteria in natural sediment microcosms. Appl Environ Microbiol 63:1531–1542
Konopka A, Turco R (1991) Biodegradation of organic compounds in vadose zone and aquifer sediments. Appl Environ Microbiol 57:2260–2268
Kuske CR, Barns SM, Busch JD (1997) Diverse uncultivated bacterial groups from soils of the arid southwestern United States that are present in many geographic regions. Appl Environ Microbiol 63:3614–3621
Kuske CR, Busch JD, Adorada DL, Dunbar JM, Barns SM (1999) Phylogeny ribosomal RNA gene typing and relative abundance of new Pseudomonas species (sensu stricto) isolated from two pinyon-juniper woodland soils of the arid southwest US. Syst Appl Microbiol 22:300–311
LaMontagne MG, Holden PA (2003) Comparison of free-living and particle-associated bacterial communities in a coastal lagoon. Microb Ecol: (in press)
LaMontagne MG, Michel FC, Holden PA, Reddy CA (2002) Evaluation of extraction and purification methods for obtaining PCR-amplifiable DNA from compost for microbial community analysis. J Microbiol Methods 49:255–264
Lane DJ, Pace B, Olsen GJ, Stahl DA, Sogin ML, Pace NR (1985) Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci USA 82:6955–6959
Lehman RM, Roberto FF, Earley D, Bruhn DF, Brink SE, O’Connell SP, Delwiche ME, Colwell FS (2001) Attached and unattached bacterial communities in a 120-meter corehole in an acidic, crystalline rock aquifer. Appl Environ Microbiol 67:2095–2106
Liu WT, Marsh TL, Cheng H, Forney LJ (1997) Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol 63:4516–4522
Ludwig W, Bauer SH, Bauer M, Held I, Kirchhof G, Schulze R, Huber I, Spring S, Hartmann A, Schleifer KH (1997) Detection and in situ identification of representatives of a widely distributed new bacterial phylum. FEMS Microbiol Lett 53:181–190
Lukow T, Dunfield PF, Liesack W (2000) Use of the T-RFLP technique to assess spatial and temporal changes in the bacterial community structure within an agricultural soil planted with transgenic and non-transgenic potato plants. FEMS Microbiol Ecol 32:241–247
Luo J, Tillman RW, White RE, Ball PR (1998) Variation in denitrification activity with soil depth under pasture. Soil Biol Biochem 30:897–903
Maidak BL, Cole JR, Lilburn TG, Parker CT, Saxman PR, Farris RJ, Garrity GM, Olsen GJ, Schmidt TM, Tiedje JM (2001) The RDP-II (Ribosomal Database Project). Nucleic Acids Res 29:173–174
McCaig AE, Glover LA, Prosser JI (2001) Numerical analysis of grassland bacterial community structure under different land management regimens by using 16S ribosomal DNA sequence data and denaturing gradient gel electrophoresis banding patterns. Appl Environ Microbiol 67:4554–4559
Moyer CL, Dobbs FC, Karl DM (1994) Estimation of diversity and community structure through restriction fragment length polymorphism distribution analysis of bacterial 16S rRNA genes from a microbial mat at an active hydrothermal vent system, Loihi Seamount, Hawaii. Appl Environ Microbiol 60:871–879
Osborn AM, Moore ERB, Timmis KN (2000) An evaluation of terminal-restriction fragment length polymorphism (T-RFLP) analysis for the study of microbial community structure and dynamics. Environ Microbiol 2:39–50
Parkin TB, Meisinger JJ (1989) Denitrification below the crop rooting zone as influenced by surface tillage. J Environ Qual 18:12–16
Polz MF, Cavanaugh CM (1998) Bias in template-to-product ratios in multitemplate PCR. Appl Environ Microbiol 64:3724–3730
Richter DD, Markewitz D (1995) How deep is soil? Bioscience 45:600–609
Rogers BF, Tate RL III (2001) Temporal analysis of the soil microbial community along a toposequence in Pineland soils. Soil Biol Biochem 33:1389–1401
Röling WFM, Milner MG, Jones DM, Lee K, Daniel F, Swannell RJP, Head IM (2002) Robust hydrocarbon degradation and dynamics of bacterial communities during nutrient-enhanced oil spill bioremediation. Appl Environ Microbiol 68:5537–5548
Suzuki MT, Giovannoni SJ (1996) Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol 62:625–630
Theron J, Cloete TE (2000) Molecular techniques for determining microbial diversity and community structure in natural environments. Crit Rev Microbiol 26:37–57
Torsvik V, Ovreas L, Thingstad TF (2002) Prokaryotic diversity—Magnitude, dynamics, and controlling factors. Science 296:1064–1066
Trumbore SE, Davidson EA, de Camargo PB, Nepstad DC, Martinelli LA (1995) Belowground cycling of carbon in forests and pastures of Eastern Amazonia. Glob Biogeochem Cy 9:515–528
Valinsky L, Vedova GD, Scupham AJ, Alvey S, Figueroa A, Yin B, Hartin RJ, Chrobak M, Crowley DE, Jiang T, Borneman J (2002) Analysis of bacterial community composition by oligonucleotide fingerprinting of rRNA genes. Appl Environ Microbiol 68:3243–3250
Westergaard K, Muller AK, Christensen S, Bloem J, Sorensen SJ (2001) Effects of tylosin as a disturbance on the soil microbial community. Soil Biol Biochem 33:2061–2071
Widmer F, Fliessbach A, Laczko E, Schulze-Aurich J, Zeyer J (2001) Assessing soil biological characteristics: a comparison of bulk soil community DNA-, PLFA-, and Biolog™-analyses. Soil Biol Biochem 33:1029–1036
Wright DH (1983) Species-energy theory: an extension of species-area theory. Oikos 41:496–506
Zhang CL, Palumbo AV, Phelps TJ, Beauchamp JJ, Brockman FJ, Murray CJ, Parsons BS, Swift DJP (1998) Grain size and depth constraints on microbial variability in coastal plain subsurface sediments. Geomicrobiol J 15:171–185
Zhou J, Bruns MA, Tiedje JM (1996) DNA recovery from soils of diverse composition. Appl Environ Microbiol 62:316–322
Zhou J, Xia B, Treves DS, Wu L-Y, Marsh TL, O’Neill RV, Palumbo AV, Tiedje JM (2002) Spatial and resource factors influencing high microbial diversity in soil. Appl Environ Microbiol 68:326–334
Author information
Authors and Affiliations
Corresponding author
Additional information
Online publication: 6 June 2003
Rights and permissions
About this article
Cite this article
LaMontagne, M.G., Schimel, J.P. & Holden, P.A. Comparison of subsurface and surface soil bacterial communities in california grassland as assessed by terminal restriction fragment length polymorphisms of PCR-amplified 16S rRNA genes. Microb Ecol 46, 216–227 (2003). https://doi.org/10.1007/s00248-003-1006-y
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/s00248-003-1006-y