Abstract
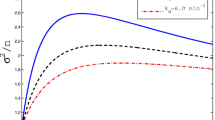
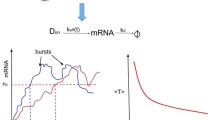
The recent in vivo RNA detection technique has allowed real-time monitoring of gene transcription in individual living cells, revealing that genes can be transcribed randomly in a bursting fashion that short periods of rapid production of multiple transcripts are interspersed with relatively long periods of no production. In this work, we utilize the three state model to study how environmental signals and the intrinsic cellular contexts are combined to regulate stochastic gene transcription. We introduce a system of three master equations to model the stochastic occurrence of transcriptional bursting. As this system cannot be solved analytically, we introduce a linear operator, called the master operator. It is of significant mathematical interests of its own and transforms the mean frequency of transcriptional bursting μ(t) and the second moment μ 2(t) into the unique solutions of the respective operator equations. Following this novel approach, we have found the exact forms of μ(t) and the variance σ 2(t). Our analysis shows that the three state transition process produces less noisy transcription than a single Poisson process does, and more transition steps average out rather than propagate fluctuations of transcripts among individual cells. The noise strength φ(t) = σ 2(t)/μ(t) displays highly non-trivial dynamics during the first two to three transcription cycles. It declines steeply from the beginning until reaching the absolute minimum value, and then bounces back suddenly to a flat level close to the steady-state. Our numerical simulations further demonstrate that the cellular signals that produce the least noisy population at steady-state may not generate the least noisy population in a finite time, and suggest that measurements at steady-state may not necessarily capture most essential features of transcription noise.
Similar content being viewed by others
References
Allen LJS (2006) Introduction to stochastic processes with biology applications, 1st edn. Prentice Hall, Englewood Cliffs
Black WJ, Kaern M, Cantor CR, Collins JJ (2003) Noise in eukaryotic gene expression. Nature 422: 633–637
Cai L, Friedman N, Xie XS (2006) Stochastic protein expression in individual cells at the single molecule level. Nature 440: 358–362
Chubb JR, Trcek T, Shenoy SM, Singer RH (2006) Transcriptional pulsing of a developmental gene. Curr Biol 16: 1018–1025
Core LJ, Lis JT (2008) Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science 319: 1791–1792
Elowitz MB, Levine AJ, Siggia ED, Swain PS (2002) Stochastic gene expression in a single cell. Science 297: 1183–1186
Golding I, Paulsson J, Zawilski SM, Cox EC (2005) Real-time kinetics of gene activity in individual bacteria. Cell 123: 1025–1036
Gorski SA, Dundr M, Misteli T (2006) The road much traveled: trafficking in the cell nucleus. Curr Opin Cell Biol 18: 284–90
Hirsch MW, Smale S, Devaney R (2003) Differential equations, dynamical systems, and an introduction to chaos, 2nd edn. Academic Press, Boston
Kaern M, Elston TC, Blake WJ, Collins JJ (2005) Stochasticity in gene expression: from theories to phenotypes. Nat Rev Genet 6: 451–464
Kaufmann BB, van Oudenaarden A (2007) Stochastic gene expression: from single molecules to the proteome. Curr Opin Genet Dev 17: 107–112
Ko MS (1991) A stochastic model for gene induction. J Theor Biol 153: 181–194
Lemon B, Tjian R (2000) Orchestrated response: a symphony of transcription factors for gene control. Genes Dev 14: 2551–2569
Maheshri N, O’Shea EK (2007) Living with noisy genes: how cells function reliably with inherent variability in gene expression. Annu Rev Biophys Biomol Struct 36: 413–434
McAdams HH, Arkin A (1997) Stochastic mechanisms in gene expression. Proc Natl Acad Sci USA 94: 814–819
Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A (2002) Regulation of noise in the expression of a single gene. Nat Genet 31: 69–73
Paulsson J (2004) Summing up the noise in gene networks. Nature 427: 415–418
Pedraza JM, Paulsson J (2008) Effects of molecular memory and bursting on fluctuations in gene expression. Science 319: 339–343
Perko L (2001) Differential equations and dynamical systems, 3rd edn. Springer, New York
Phair RD, Misteli T (2000) High mobility of proteins in the mammalian cell nucleus. Nature 04: 604–609
Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S (2006) Stochastic mRNA synthesis in mammalian cells. PLoS Biol 4: 1707–1719
Raser JM, O’Shea EK (2004) Control of stochasticity in eukaryotic gene expression. Science 304: 1811–1814
Reppas NB, Wade JT, Church GM, Struhl K (2006) The transition between transcriptional initiation and elongation in E. coli is often rate-limiting, variable, and associated with rapid release of σ70. Mol Cell 24: 747–757
Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB (2005) Gene regulation at the single-cell level. Science 307: 1962–1965
Schrodinger E (1944) What is life? The physical aspect of the living cell. Cambridge University Press, Cambridge
Swain PS, Elowitz MB, Siggia ED (2002) Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc Natl Acad Sci USA 99: 12795–12800
Tang M (2008) The mean and noise of stochastic gene transcription. J Theor Biol 253: 271–280
Thattai M, van Oudenaarden A (2001) Intrinsic noise in gene regulatory networks. Proc Natl Acad Sci USA 98: 8614–8619
van Kampen NG (2007) Stochastic processes in physics and chemistry, 3rd edn. North-Holland, Amsterdam
Wade JT, Struhl K (2007) The transition from transcription initiation to elongation. Curr Opin Genet Dev 18: 1–7
Yu J, Xiao J, Ren X, Lao K, Xie XS (2006) Probing gene expression in live cells, one protein molecule at a time. Science 311: 1600–1603
Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA (2007) RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet 39: 1512–1516
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Tang, M. The mean frequency of transcriptional bursting and its variation in single cells. J. Math. Biol. 60, 27–58 (2010). https://doi.org/10.1007/s00285-009-0258-7
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00285-009-0258-7