Abstract
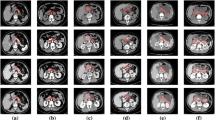
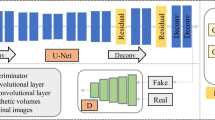
Organ segmentation from existing imaging is vital to the medical image analysis and disease diagnosis. However, the boundary shapes and area sizes of the target region tend to be diverse and flexible. And the frequent applications of pooling operations in traditional segmentor result in the loss of spatial information which is advantageous to segmentation. All these issues pose challenges and difficulties for accurate organ segmentation from medical imaging, particularly for organs with small volumes and variable shapes such as the pancreas. To offset aforesaid information loss, we propose a deep convolutional neural network (DCNN) named multi-scale selection and multi-channel fusion segmentation model (MSC-DUnet) for pancreas segmentation. This proposed model contains three stages to collect detailed cues for accurate segmentation: (1) increasing the consistency between the distributions of the output probability maps from the segmentor and the original samples by involving the adversarial mechanism that can capture spatial distributions, (2) gathering global spatial features from several receptive fields via multi-scale field selection (MSFS), and (3) integrating multi-level features located in varying network positions through the multi-channel fusion module (MCFM). Experimental results on the NIH Pancreas-CT dataset show that our proposed MSC-DUnet obtains superior performance to the baseline network by achieving an improvement of 5.1% in index dice similarity coefficient (DSC), which adequately indicates that MSC-DUnet has great potential for pancreas segmentation.










Similar content being viewed by others
References
E. Gibson, F. Giganti, Y. Hu, E. Bonmati, S. Bandula, K. Gurusamy, B. Davidson, S.P. Pereira, M.J. Clarkson, D.C.J.I.t.o.m.i. Barratt, Automatic multi-organ segmentation on abdominal CT with dense v-networks, IEEE transactions on medical imaging, 37 (2018) 1822–1834.
A. Balagopal, S. Kazemifar, D. Nguyen, M.-H. Lin, R. Hannan, A. Owrangi, S.J.P.i.M. Jiang, Biology, Fully automated organ segmentation in male pelvic CT images, Physics in Medicine & Biology, 63 (2018) 245015.
M. Anthimopoulos, S. Christodoulidis, L. Ebner, A. Christe, S.J.I.t.o.m.i. Mougiakakou, Lung pattern classification for interstitial lung diseases using a deep convolutional neural network, IEEE transactions on medical imaging, 35 (2016) 1207–1216.
J. Xu, X. Luo, G. Wang, H. Gilmore, A.J.N. Madabhushi, A deep convolutional neural network for segmenting and classifying epithelial and stromal regions in histopathological images, Neurocomputing, 191 (2016) 214-223.
T. Mordan, N. Thome, G. Henaff, M. Cord, Revisiting multi-task learning with rock: a deep residual auxiliary block for visual detection, Advances in Neural Information Processing Systems, 2018, pp. 1310–1322.
L. Abel, J. Wasserthal, T. Weikert, A.W. Sauter, I. Nesic, M. Obradovic, S. Yang, S. Manneck, C. Glessgen, J.M. Ospel, B. Stieltjes, D.T. Boll, B. Friebe, Automated detection of pancreatic cystic lesions on CT using deep learning, Diagnostics, 11 (2021) 901.
J. Yue-Hei Ng, F. Yang, L.S. Davis, Exploiting local features from deep networks for image retrieval, Proceedings of the IEEE conference on computer vision and pattern recognition workshops, 2015, pp. 53–61.
V. Badrinarayanan, A. Kendall, R.J.I.t.o.p.a. Cipolla, m. intelligence, Segnet: a deep convolutional encoder-decoder architecture for image segmentation, IEEE transactions on pattern analysis and machine intelligence, 39 (2017) 2481–2495.
L. Liu, F.-X. Wu, J.J.N. Wang, Efficient multi-kernel DCNN with pixel dropout for stroke MRI segmentation, Neurocomputing, 350 (2019) 117-127.
J. Long, E. Shelhamer, T. Darrell, Fully convolutional networks for semantic segmentation, Proceedings of the IEEE conference on computer vision and pattern recognition, 2015, pp. 3431–3440.
H.R. Roth, A. Farag, E. Turkbey, L. Lu, J. Liu, R.M. Summers, data from pancreas-CT. The cancer imaging archive, ed, 2016.
K. Clark, B. Vendt, K. Smith, J. Freymann, J. Kirby, P. Koppel, S. Moore, S. Phillips, D. Maffitt, M.J.J.o.d.i. Pringle, The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository, Journal of digital imaging, 26 (2013) 1045–1057.
X. Li, Q. Dou, H. Chen, C.-W. Fu, X. Qi, D.L. Belavý, G. Armbrecht, D. Felsenberg, G. Zheng, P.-A.J.M.i.a. Heng, 3D multi-scale FCN with random modality voxel dropout learning for intervertebral disc localization and segmentation from multi-modality MR images, Medical image analysis, 45 (2018) 41–54.
H.R. Roth, H. Oda, X. Zhou, N. Shimizu, Y. Yang, Y. Hayashi, M. Oda, M. Fujiwara, K. Misawa, K.J.C.M.I. Mori, Graphics, An application of cascaded 3D fully convolutional networks for medical image segmentation, Computerized Medical Imaging and Graphics, 66 (2018) 90–99.
X. Zhou, R. Takayama, S. Wang, T. Hara, H.J.M.p. Fujita, Deep learning of the sectional appearances of 3D CT images for anatomical structure segmentation based on an FCN voting method, Medical physics, 44 (2017) 5221–5233.
O. Ronneberger, P. Fischer, T. Brox, U-net: Convolutional networks for biomedical image segmentation, International Conference on Medical image computing and computer-assisted intervention, Springer, 2015, pp. 234–241.
Z. Zhong, Y. Kim, K. Plichta, B.G. Allen, L. Zhou, J. Buatti, X.J.M.p. Wu, Simultaneous cosegmentation of tumors in PET‐CT images using deep fully convolutional networks, Medical physics, 46 (2019) 619–633.
F. Isensee, P.F. Jaeger, P.M. Full, I. Wolf, S. Engelhardt, K.H. Maier-Hein, Automatic cardiac disease assessment on cine-MRI via time-series segmentation and domain specific features, International workshop on statistical atlases and computational models of the heart, Springer, 2017, pp. 120-129.
Z. Meng, Z. Fan, Z. Zhao, F. Su, ENS-Unet: End-to-end noise suppression U-Net for brain tumor segmentation, 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), IEEE, 2018, pp. 5886-5889.
I. Goodfellow, J. Pouget-Abadie, M. Mirza, B. Xu, D. Warde-Farley, S. Ozair, A. Courville, Y. Bengio, Generative adversarial nets, Advances in neural information processing systems, 2014, pp. 2672–2680.
P. Luc, C. Couprie, S. Chintala, J. Verbeek, Semantic segmentation using adversarial networks, arXiv preprint, https://arxiv.org/abs/1611.08408 (2016).
S. Nema, A. Dudhane, S. Murala, S.J.B.S.P. Naidu, Control, RescueNet: an unpaired GAN for brain tumor segmentation, Biomedical Signal Processing and Control, 55 (2020) 101641.
X. Dong, Y. Lei, T. Wang, M. Thomas, L. Tang, W.J. Curran, T. Liu, X.J.M.p. Yang, Automatic multiorgan segmentation in thorax CT images using U‐net‐GAN, Medical physics, 46 (2019) 2157–2168.
A. Paszke, S. Gross, S. Chintala, G. Chanan, E. Yang, Z. DeVito, Z. Lin, A. Desmaison, L. Antiga, A. Lerer, Automatic differentiation in PyTorch, NIPS Autodiff workshop (2017).
H.R. Roth, L. Lu, N. Lay, Spatial aggregation of holistically-nested convolutional neural networks for automated pancreas localization and segmentation, Medical image analysis, 45 (2018) 94-107.
Y. Zhou, L. Xie, W. Shen, Y. Wang, E.K. Fishman, A.L. Yuille, A fixed-point model for pancreas segmentation in abdominal CT scans, International conference on medical image computing and computer-assisted intervention, Springer, 2017, pp. 693-701.
J. Cai, L. Lu, Y. Xie, Improving deep pancreas segmentation in CT and MRI images via recurrent neural contextual learning and direct loss function, MICCAI, 2017.
Q. Yu, L. Xie, Y. Wang, Y. Zhou, Recurrent saliency transformation network: Incorporating multi-stage visual cues for small organ segmentation, Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2018, pp. 8280–8289.
J. Cai, L. Lu, F. Xing, Pancreas segmentation in CT and MRI images via domain specific network designing and recurrent neural contextual learning, arXiv preprint, https://arxiv.org/abs/1803.11303 (2018).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Li, M., Lian, F. & Guo, S. Multi-scale Selection and Multi-channel Fusion Model for Pancreas Segmentation Using Adversarial Deep Convolutional Nets. J Digit Imaging 35, 47–55 (2022). https://doi.org/10.1007/s10278-021-00563-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-021-00563-x