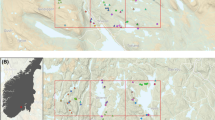
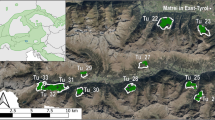
Abstract
Estimating the size of animal populations is essential for understanding the demography and conservation status of species. Genetic Non-Invasive Sampling (gNIS) combined with Spatially Explicit Capture-Recapture (SECR) modelling may provide a practical tool to obtain such estimates. Here, we evaluate for the first time the potential and limitations of this approach to estimate population densities for small mammals inhabiting patchily distributed habitats, focusing on the endemic Iberian Cabrera vole (Microtus cabrerae). Using 11 highly polymorphic microsatellites and two sex-linked introns, we compared population estimates in November/December 2011 based on live-trapping and gNIS and assessed the impact of distinct consensus criteria to differentiate unique genotypes. Live-trapping over 21 days captured 31 individuals, while gNIS over 5 days recorded 65–69 individuals. SECR models indicated that individual detectability was positively affected by live-trapping capture success on the previous occasion, while for gNIS, it was mainly affected by genotyping success rates and patch size. Live-trapping produced the lowest density estimates (mean ± SE) of 16.6 ± 3.2 individuals per hectare of suitable habitat (ind/ha). Estimates based on gNIS were higher and varied slightly between 25.2 ± 4.0 and 28.8 ± 4.5 ind/ha depending on assuming one or two genotyping errors, respectively, when differentiating individual genetic profiles. Results suggest that live-trapping underestimated the vole population, while the larger number of individuals detected through gNIS allowed better estimates with lower field effort. Overall, we suggest that gNIS combined with SECR models provides an effective tool to estimate small mammal population densities in fragmented habitats.

Similar content being viewed by others
References
AEMET, IM (2011) Atlas climático Ibérico / Iberian climate atlas. Agencia Estatal de Meteorología, Ministerio de Medio Ambiente y Rural y Marino, Madrid. Instituto de Meteorologia de Portugal
Alasaad S, Sánchez A, Marchal JA, Píriz A, Garrido-García JA, Carro F, Romero I, Soriguer RC (2011) Efficient identification of Microtus cabrerae excrements using noninvasive molecular analysis. Conserv Genet Resour 3:127–129. https://doi.org/10.1007/s12686-010-9306-2
Arandjelovic M, Vigilant L (2018) Non-invasive genetic censusing and monitoring of primate populations. Am J Primatol 80:e22743. https://doi.org/10.1002/ajp.22743
Barbosa S, Pauperio J, Searle JB, Alves PC (2013) Genetic identification of Iberian rodent species using both mitochondrial and nuclear loci: application to noninvasive sampling. Mol Ecol Resour 13:43–56. https://doi.org/10.1111/1755-0998.12024
Beja-Pereira A, Oliveira R, Alves PC, Schwartz MK, Luikart G (2009) Advancing ecological understandings through technological transformations in noninvasive genetics. Mol Ecol Resour 9:1279–1301. https://doi.org/10.1111/j.1755-0998.2009.02699.x
Bonin A, Bellemain E, Bronken Eidesen P, Pompanon F, Brochmann C, Taberlet P (2004) How to track and assess genotyping errors in population genetics studies. Mol Ecol 13:3261–3273. https://doi.org/10.1111/j.1365-294X.2004.02346.x
Borchers DL, Efford MG (2008) Spatially explicit maximum likelihood methods for capture-recapture studies. Biometrics 64:377–385. https://doi.org/10.1111/j.1541-0420.2007.00927.x
Bowers MA, Matter SF (1997) Landscape ecology of mammals: relationships between density and patch-size. J Mammal 78:999–1013. https://doi.org/10.2307/1383044
Brazeal JL, Weist T, Sacks BN (2017) Noninvasive genetic spatial capture-recapture for estimating deer population abundance. J Wildl Manag 81:629–640. https://doi.org/10.1002/jwmg.21236
Brinkman TJ, Schwartz MK, Person DK, Pilgrim KL, Hundertmark KJ (2010) Effects of time and rainfall on PCR success using DNA extracted from deer fecal pellets. Conserv Genet 11:1547–1552. https://doi.org/10.1007/s10592-009-9928-7
Burnham KP, Anderson DR (2002) Model selection and multimodel inference: a practical information-theoretic approach, 2nd edn. Springer-Verlag, New York
Cheng E, Hodges KE, Sollmann R, Mills LS (2017) Genetic sampling for estimating density of common species. Ecol Evol 7:6210–6219. https://doi.org/10.1002/ece3.3137
Conroy MJ, Runge JP, Barker RJ, Schofield MR, Fonnesbeck CJ (2008) Efficient estimation of abundance for patchily distributed populations via two-phase, adaptive sampling. Ecology 89:3362–3370. https://doi.org/10.1890/07-2145.1
De Barba M, Miquel C, Lobréaux S, Quenette PY, Swenson JE, Taberlet P (2017) High-throughput microsatellite genotyping in ecology: improved accuracy, efficiency, standardization and success with low-quantity and degraded DNA. Mol Ecol Resour 17:492–507. https://doi.org/10.1111/1755-0998.12594
Do R, Shonfield J, McAdam AG (2013) Reducing accidental shrew mortality associated with small-mammal livetrapping II: a field experiment with bait supplementation. J Mammal 94:754–760. https://doi.org/10.1644/12-MAMM-A-242.1
Efford M (2004) Density estimation in live-trapping studies. Oikos 106:598–610. https://doi.org/10.1111/j.0030-1299.2004.13043.x
Efford MG (2011) Estimation of population density by spatially explicit capture-recapture analysis of data from area searches. Ecology 92:2202–2207. https://doi.org/10.1890/11-0332.1
Efford M (2014) secr: Spatially explicit capture-recapture models. R package version 2.8.2 http://www.otago.ac.nz/density. Accessed 30 April 2014
Efford M (2018) Polygon and transect detectors in secr 3.1. http://www.otago.ac.nz/density/pdfs/secr-polygondetectors.pdf. Accessed 16 March 2018
Efford MG, Fewster RM (2013) Estimating population size by spatially explicit capture-recapture. Oikos 122:918–928. https://doi.org/10.1111/j.1600-0706.2012.20440.x
Efford MG, Borchers DL, Byrom AE (2009) Density estimation by spatially explicit capture–recapture: likelihood-based methods. In: Thomson DL, Cooch EG, Conroy MJ (eds) Modeling demographic processes in marked populations. Springer, Boston, pp 255–269. https://doi.org/10.1007/978-0-387-78151-8_11
Fernández-Salvador R, Ventura J, García-Perea R (2005) Breeding patterns and demography of a population of the Cabrera vole, Microtus cabrerae. Anim Biol 55:147–161. https://doi.org/10.1163/1570756053993497
Ferreira CM, Sabino-Marques H, Paupério J, Barbosa S, Costa P, Encarnação C, Alpizar-Jara R, Pita R, Beja P, Mira A, Searle JB, Alves PC (2018) Genetic non-invasive sampling (gNIS) as a cost-effective tool for monitoring elusive small mammals. Eur J Wildl Res in-press. https://doi.org/10.1007/s10344-018-1188-8
Fletcher QE, Boonstra R (2006) Impact of live trapping on the stress response of the meadow vole (Microtus pennsylvanicus). J Zool 270:473–478. https://doi.org/10.1111/j.1469-7998.2006.00153.x
Garrido-García JA, Soriguer RC (2014) Topillo de Cabrera Iberomys cabrerae (Thomas, 1906) In: Calzada J, Clavero M, Fernández A. (eds). Guía virtual de los indicios de los mamíferos de la Península Ibérica, Islas Baleares y Canarias. Sociedad Española para la Conservación y Estudio de los Mamíferos (SECEM). http://www.secem.es/guiadeindiciosmamiferos/. Accessed 20 April 2016
Johannesen E, Andreassen HP, Ims RA (2000) Spatial explicit demography: the effects habitat patch isolation have on vole matrilines. Ecol Lett 3:48–57. https://doi.org/10.1046/j.1461-0248.2000.00119.x
Johnson PCD, Haydon DT (2007) Maximum-likelihood estimation of allelic dropout and false allele error rates from microsatellite genotypes in the absence of reference data. Genetics 175:827–842. https://doi.org/10.1534/genetics.106.064618
Kalinowski ST, Sawaya MA, Taper ML (2006) Individual identification and distribution of genotypic differences between individuals. J Wildl Manag 70:1148–1150. https://doi.org/10.2193/0022-541X(2006)70[1148:IIADOG]2.0.CO;2
Kéry M, Gardner B, Stoeckle T, Weber D, Royle JA (2010) Use of spatial capture–recapture modeling and DNA data to estimate densities of elusive animals. Conserv Biol 25:356–364. https://doi.org/10.1111/j.1523-1739.2010.01616.x
Lampa S, Henle K, Klenke R, Hoehn M, Gruber B (2013) How to overcome genotyping errors in non-invasive genetic mark-recapture population size estimation—a review of available methods illustrated by a case study. J Wildl Manag 77:1490–1511. https://doi.org/10.1002/jwmg.604
Landete-Castillejos T, Andrés-Abellán M, Argandoña JJ, Garde J (2000) Distribution of the Cabrera vole in its first reported areas reassessed by live-trapping. Biol Conserv 94:127–130. https://doi.org/10.1016/S0006-3207(99)00167-6
López-Bao JV, Godinho R, Pacheco C, Lema FJ, García E, Llaneza L, Palacios V, Jiménez J (2018) Toward reliable population estimates of wolves by combining spatial capture-recapture models and non-invasive DNA monitoring. Sci Rep 8:2177. https://doi.org/10.1038/s41598-018-20675-9
Lounsberry ZT, Forrester TD, Olegario MT, Brazeal JL, Wittmer HU, Sacks BN (2015) Estimating sex-specific abundance in fawning areas of a high-density Columbian black-tailed deer population using fecal DNA. J Wildl Manag 79:39–49. https://doi.org/10.1002/jwmg.817
Luikart G, Ryman N, Tallmon DA, Schwartz MK, Allendorf FW (2010) Estimation of census and effective population sizes: the increasing usefulness of DNA-based approaches. Conserv Genet 11:355–373. https://doi.org/10.1007/s10592-010-0050-7
Lukacs PM, Burnham KP (2005) Review of capture-recapture methods applicable to noninvasive genetic sampling. Mol Ecol 14:3909–3919. https://doi.org/10.1111/j.1365-294X.2005.02717.x
Macbeth GM, Broderick D, Ovenden JR, Buckworth RC (2011) Likelihood-based genetic mark–recapture estimates when genotype samples are incomplete and contain typing errors. Theor Popul Biol 80:185–196. https://doi.org/10.1016/j.tpb.2011.06.006
MacGregor-Fors I, Payton ME (2013) Contrasting diversity values: statistical inferences based on overlapping confidence intervals. PLoS One 8:e56794. https://doi.org/10.1371/journal.pone.0056794
Manly BFJ (2004) Two-phase adaptive stratified sampling. In: Thompson WL (ed) Sampling rare or elusive species: concepts, designs, and techniques for estimating population parameters. Island Press, Washington DC, pp 123–133
McCravy KW, Rose RK (1992) An analysis of external features as predictors of reproductive status in small mammals. J Mammal 73:151–159. https://doi.org/10.2307/1381877
McKelvey KS, Schwartz MK (2004) Genetic errors associated with population estimation using non-invasive molecular tagging: problems and new solutions. J Wildl Manag 68:439–448. https://doi.org/10.2193/0022-541X(2004)068[0439,GEAWPE]2.0.CO;2
Mills LS, Citta JJ, Lair KP, Schwarz MK, Tallman DA (2000) Estimating animal abundance using noninvasive DNA sampling: promise and pitfalls. Ecol Appl 10:283–294. https://doi.org/10.1890/1051-0761(2000)010[0283:EAAUND]2.0.CO;2
Mollet P, Kéry M, Gardner B, Pasinelli G, Royle JA (2015) Estimating population size for capercaillie (Tetrao urogallus L.) with spatial capture-recapture models based on genotypes from one field sample. PLoS One 10:e0129020. https://doi.org/10.1371/journal.pone.0129020
Mondol S, Karanth KU, Kumar NS, Gopalaswamy AM, Andheria A, Ramakrishnan U (2009) Evaluation of non-invasive genetic sampling methods for estimating tiger population size. Biol Conserv 142:2350–2360. https://doi.org/10.1016/j.biocon.2009.05.014
Morin DJ, Waits LP, McNitt DC, Kelly MJ (2018) Efficient single-survey estimation of carnivore density using fecal DNA and spatial capture-recapture: a bobcat case study. Popul Ecol 60:197–209. https://doi.org/10.1007/s10144-018-0606-9
Murphy MA, Kendall KC, Robinson A, Waits LP (2007) The impact of time and field conditions on brown bear (Ursus arctos) faecal DNA amplification. Conserv Genet 8:1219–1224. https://doi.org/10.1007/s10592-006-9264-0
Murphy SM, Augustine BC, Ulrey WA, Guthrie JM, Scheick BK, McCown JW, Cox JJ (2017) Consequences of severe habitat fragmentation on density, genetics, and spatial capture-recapture analysis of a small bear population. PLoS One 12:e0181849. https://doi.org/10.1371/journal.pone.0181849
Okello JBA, Wittemyer G, Rasmussen HB, Douglas-Hamilton I, Nyakaana S, Arctander P, Siegismund HR (2005) Noninvasive genotyping and Mendelian analysis of microsatellites in African savannah elephants. J Hered 96:679–687. https://doi.org/10.1093/jhered/esi117
Piñero FS, Garrido-García JA, Soriguer RC (2012) Dung beetles (Scarabaeidae, Coleoptera) of latrines of the Iberian endemic rodent Microtus cabrerae (Rodentia: Cricetidae: Microtinae) at Sierra de Segura (S. Iberian Peninsula). Bol Asoc Esp Entomol 36:451–455
Pita R, Beja P, Mira A (2007) Spatial population structure of the Cabrera vole in Mediterranean farmland: the relative role of patch and matrix effects. Biol Conserv 134:383–392. https://doi.org/10.1016/j.biocon.2006.08.026
Pita R, Mira A, Beja P (2010) Spatial segregation of two vole species (Arvicola sapidus and Microtus cabrerae) within habitat patches in a highly fragmented farmland landscape. Eur J Wildl Res 56:651–662. https://doi.org/10.1007/s10344-009-0360-6
Pita R, Mira A, Beja P (2011) Assessing habitat differentiation between coexisting species: the role of spatial scale. Acta Oecol 37:124–132. https://doi.org/10.1016/j.actao.2011.01.006
Pita R, Mira A, Beja P (2014) Microtus cabrerae (Rodentia: Cricetidae). Mamm Species 46(912):48–70. https://doi.org/10.1644/912.1
Pita R, Lambin X, Mira A, Beja P (2016) Hierarchical spatial segregation of two Mediterranean vole species: the role of patch-network structure and matrix composition. Oecologia 182:253–263. https://doi.org/10.1007/s00442-016-3653-y
R Development Core Team (2014) R: a language and environment for statistical computing, 3.0. R Foundation for Statistical Computing, Vienna, p 2 http://www.R-project.org. Accessed 30 April 2014
Rehnus M, Bollman K (2016) Non-invasive genetic population density estimation of mountain hares (Lepus timidus) in the Alps: systematic or opportunistic sampling? Eur J Wildl Res 62:737–747. https://doi.org/10.1007/s10344-016-1053-6
Rodgers TW, Giacalone J, Heske EJ, Janečka JE, Phillips CA, Schooley RL (2014) Comparison of noninvasive genetics and camera trapping for estimating population density of ocelots (Leopardus pardalis) on Barro Colorado Island, Panama. Trop Conserv Sci 7:690–705. https://doi.org/10.1177/194008291400700408
Rosário IT (2012) Towards a conservation strategy for an endangered rodent, the Cabrera vole (Microtus cabrerae Thomas): insights from ecological data. Ph.D. Dissertation, University of Lisbon
Royle JA, Chandler RB, Sollmann R, Gardner B (2014) Spatial capture-recapture. Academic Press, Waltham
Royle JA, Fuller AK, Sutherland C (2018) Unifying population and landscape ecology with spatial capture–recapture. Ecography 41:444–456. https://doi.org/10.1111/ecog.03170
Santini A, Lucchini V, Fabbri E, Randi E (2007) Ageing and environmental factors affect PCR success in wolf (Canis lupus) excremental DNA samples. Mol Ecol Notes 7:955–961. https://doi.org/10.1111/j.1471-8286.2007.01829.x
Scharine PD, Nielsen CK, Schauber EM, Rubert L, Crawford JC (2011) Occupancy, detection, and habitat associations of sympatric lagomorphs in early-successional bottomland forests. J Mammal 92:880–890. https://doi.org/10.1644/10-MAMM-A-078.1
Schwartz MK, Luikart G, Waples RS (2007) Genetic monitoring as a promising tool for conservation and management. Trends Ecol Evol 22:25–33. https://doi.org/10.1016/j.tree.2006.08.009
Sikes RS, Gannon WL, the Animal Care and Use Committee of the American Society of Mammalogists (2011) Guidelines of the American Society of Mammalogists for the use of wild mammals in research. J Mammal 92:235–253. https://doi.org/10.1644/10-MAMM-F-355.1
Slauson KM, Zielinski WJ, Schwartz MK (2017) Ski areas affect Pacific marten movement, habitat use, and density. J Wildl Manag 81:892–904. https://doi.org/10.1002/jwmg.21243
Solberg KH, Bellemain E, Drageset OM, Taberlet P, Swenson JE (2006) An evaluation of field and non-invasive genetic methods to estimate brown bear (Ursus arctos) population size. Biol Conserv 128:158–168. https://doi.org/10.1016/j.biocon.2005.09.025
Stem C, Margoluis R, Salafsky N, Brown M (2005) Monitoring and evaluation in conservation: a review of trends and approaches. Conserv Biol 19:295–309. https://doi.org/10.1111/j.1523-1739.2005.00594.x
Taberlet P, Waits LP, Luikart G (1999) Noninvasive genetic sampling: look before you leap. Trends Ecol Evol 14:323–327. https://doi.org/10.1016/S0169-5347(99)01637-7
Thompson WL (ed) (2004) Sampling rare or elusive species: concepts, designs, and techniques for estimating population parameters. Island Press, Washington DC
Thompson CM, Royle JA, Garner JD (2012) A framework for inference about carnivore density from unstructured spatial sampling of scat using detector dogs. J Wildl Manag 76:863–871. https://doi.org/10.1002/jwmg.317
Traill LW, Bradshaw CJA, Brook BW (2007) Minimum viable population size: a meta-analysis of 30 years of published estimates. Biol Conserv 139:159–166. https://doi.org/10.1016/j.biocon.2007.06.011
Turner MG, Gardner RH, O’Neill RV (2001) Landscape ecology in theory and practice: pattern and process. Springer-Verlag, New York
Valière N (2002) GIMLET: a computer program for analysing genetic individual identification data. Mol Ecol Notes 2:377–379. https://doi.org/10.1046/j.1471-8286.2002.00228.x-i2
Waits JL, Leberg PL (2000) Biases associated with population estimation using molecular tagging. Anim Conserv 3:191–199. https://doi.org/10.1111/j.1469-1795.2000.tb00103.x
Waits LP, Luikart G, Taberlet P (2001) Estimating the probability of identity among genotypes in natural populations: cautions and guidelines. Mol Ecol 10:249–256. https://doi.org/10.1046/j.1365-294X.2001.01185.x
Acknowledgments
This study was funded by FEDER through the Programa Operacional Factores de Competitividade—COMPETE and the Portuguese Foundation for Science and Technology—FCT—within the scope of the projects PERSIST (PTDC/BIA-BEC/105110/2008), NETPERSIST (PTDC/AAG-MAA/3227/2012) and MateFrag (PTDC/BIA-BIC/6582/2014). HSM was supported by the FCT grant SFRH/BD/73765/2010. JP was supported by a postdoctoral grant funded by the project ‘Genomics and Evolutionary Biology’ co-financed by North Portugal Regional Operational Programme 2007/2013 (ON.2 - O Novo Norte), under the National Strategic Reference Framework, through the ERDF. PB was supported by EDP Biodiversity Chair. RP was supported by the FCT grants SFRH/BPD/73478/2010 and SFRH/BPD/109235/2015. We thank Estrela Matilde for her assistance in fieldwork. We are grateful to Murray Efford for guidance through the secr package and, together with Tiago Marques, for advice regarding data analysis. Ana Galantinho provided useful feedback on a previous version of the manuscript. We also thank two anonymous reviewers who provided valuable suggestions to improve the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All applicable international, national and/or institutional guidelines for the care and use of animals were followed. All procedures were carried out under permission from the Portuguese biodiversity conservation agency (ICNF—Instituto de Conservação da Natureza e das Florestas, permit nos. 76, 77 and 80/2011/CAPT) and conformed to the guidelines approved by the American Society of Mammalogists (Sikes et al. 2011).
Electronic supplementary material
ESM 1
(PDF 369 kb)
Rights and permissions
About this article
Cite this article
Sabino-Marques, H., Ferreira, C.M., Paupério, J. et al. Combining genetic non-invasive sampling with spatially explicit capture-recapture models for density estimation of a patchily distributed small mammal. Eur J Wildl Res 64, 44 (2018). https://doi.org/10.1007/s10344-018-1206-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10344-018-1206-x