Abstract
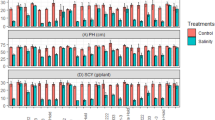
Soil salinity reduces cotton growth, yield, and fiber quality and has become a serious problem in the arid southwestern region of the Unites States. Development and planting of salt-tolerant cultivars could ameliorate the deleterious effects. The objective of this study was to assess the genetic variation of salt tolerance and identify salt tolerant genotypes in a backcross inbred line (BIL) population of 142 lines from a cross of Upland (Gossypium hirsutum) × Pima cotton (G. barbadense) at the seedling growth stage. As compared with the non-saline (control) conditions, seedlings under the salinity stress (200 mM NaCl) showed a significant reduction in all the plant growth characteristics measured, as expected. Even though the two parents did not differ in salt response as measured by percent reduction, significant genotype variations in the BIL population were detected for all traits except for leaf number. Based on percent reduction of the traits measured, several BILs were more salt tolerant than both parents. The results indicate that transgressive segregation occurred during the process of backcrossing and selfing even though both parents were not salt tolerant during seedling growth. Coefficients of correlation between all the traits were significantly positive, indicating an association between the traits measured. The estimates of broad-sense heritability were 0.69, 0.46, 0.47, 0.43, and 0.49 for plant height, fresh weight of shoot and root, and dry weight of shoot and root, respectively, indicating that salt tolerance during cotton seedling growth is moderately heritable and environmental variation plays an equally important role. The overall results demonstrate that backcrossing followed by repeated self-pollination is a successful strategy to enhance salt tolerance at the seedling stage by transferring genetic factors from Pima to Upland cotton.




Similar content being viewed by others
References
Abbas G, Khan TM, Khan AA, Khan AI (2011) Discrimination of salt tolerant and susceptible cotton genotypes at seedling stage using selection index. Int J Agric Biol 13:339–345
Abdelraheem A, Tiwari R, Zhang JF (2012) Genetic analysis and QTL mapping of drought tolerance in cotton under PEG conditions. Proc. Beltwide Cotton Conf. pp 719–728
Abul-Naas AA, Omran MS (1974) Salt tolerance of seventeen cotton cultivars during germination and early seedling development. Z Acker und Pflanzenbau 140:229–236
Ashraf M, Ahmad S (1999) Exploitation of intra-specific genetic variation for improvement of salt (NaCl) tolerance in Upland cotton (Gossypium hirsutum L.). Hereditas 131:253–256
Ayers AD, Brown JW, Wadleigh CH (1952) Salt tolerance of barley and wheat in soil plots receiving several salinization regimes. Agron J 44:307–310
Basal H, Hemphill JK, Smith CW (2006) Shoot and root characteristics of converted race stocks accessions of upland cotton (Gossypium hirsutum L.) grown under salt stress conditions. Am J Plant Physiol 1:99–106
Bhatti MA, Azhar FM (2002) Salt tolerance of nine Gossypium hirsutum L. varieties to NaCl salinity at early stage of plant development. Int J Agric Biol 4:544–546
Chee P, Draye X, Jiang CX, Decanini L, Delmonte TA, Bredhauer R, Smith CW, Paterson AH (2005) Molecular dissection of interspecific variation between Gossypium hirsutum and Gossypium barbadense (cotton) by a backcross-self approach: I. Fiber elongation. Theor Appl Genet 111:757–763
Draye X, Chee P, Jiang CX, Decanini L, Delmonte TA, Bredhauer R, Smith CW, Paterson AH (2005) Molecular dissection of interspecific variation between Gossypium hirsutum and G. barbadense (cotton) by a backcross-self approach: II. Fiber fineness. Theor Appl Genet 111:764–771
FAO (2008) FAO: Land and plant nutrition management service. http://www.fao.org/ag/agl/agll/spush. Accessed 24 April 2013
Gorham J, Läuchli A, Leidi EO (2010) Plant responses to salinity. In: Stewart JM, Oosterhuis DM, Heitholt JJ, Mauney JR (eds) Physiology of cotton. Springer, Netherlands, pp 129–141
Hanif M, Noor E, Murtaza N, Qayyum A, Malik W (2008) Assessment of variability for salt tolerance at seedling stage in Gossypium hirsutum L. J Food Agric Environ 6:134–138
Higbie SM, Wang F, Stewart JM, Sterling TM, Lindemann WC, Hughs E, Zhang JF (2010) Physiological response to salt (NaCl) stress in selected cultivated tetraploid cottons. Int J Agron 2010:1–12
Holland JB, Nyquist WE, Cervantes-Martínez CT (2002) Estimating and interpreting heritability for plant breeding: an update. Plant Breed Rev 22:9–112
Lande R (1981) The minimum number of genes contributing to quantitative variation between and within populations. Genetics 99:541–553
Läuchli A, Grattan SR (2007) Plant growth and development under salinity stress. In: Jenks MA, Hasegawa PM, Jain SM (eds) Advances in molecular breeding toward drought and salt tolerant crops. Springer, Netherlands, pp 1–32
Lobell DB, Ortiz-Monasterio JI, Gurrola FC, Valenzuela L (2007) Identification of saline soils with multiyear remote sensing of crop yields. Soil Sci Soc Am J 71:777–783
Maas EV, Hoffman GJ (1977) Crop salt tolerance-current assessment. J Irrigation Drainage Div, ASCE 103:115–134
Maas EV, Hoffman GJ, Chaba GD, Poss JA, Shannon MC (1983) Salt sensitivity of corn at various growth stages. Irrigation Sci 4:45–57
Mano Y, Takeda K (1997) Mapping quantitative trait loci for salt tolerance at germination and the seedling stage in barley (Hordeum vulgare L.). Euphytica 94:263–272
Mer RK, Prajith PK, Pandya DH, Pandey AN (2000) Effect of salts on germination of seeds and growth of young plants of Hordeum vulgare, Triticum aestivum, Cicer arietinum and Brassica juncea. J Agron Crop Sci 185:209–217
Munns R (2002) Comparative physiology of salt and water stress. Plant Cell Environ 25:239–250
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Ann Rev Plant Biol 59:651–681
Munns R, Schachtman DP, Condon AG (1995) The significance of a two-phase growth-response to salinity in wheat and barley. Aust J Plant Physiol 22:561–569
Munns R, James RA, Lauchli A (2006) Approaches to increasing the salt tolerance of wheat and other cereals. J Exp Bot 57:1025–1043
Pang MX, Percy RG, Hughs SE, Jones DC, Zhang JF (2012) Identification of genes that were differentially expressed and associated with fiber yield and quality using cDNA-AFLP and a backcross inbred line population. Mol Breed 30:975–985
Pearson GA, Ayers AD, Eberhard DL (1966) Relative salt tolerance of rice during germination and early seedling development. Soil Sci 102:151–156
Pitman MG, Lauchli A (2002) Global impact of salinity and agricultural ecosystyems. In: Läuchli A, Lüttge U (eds) Salinity: environment-plants-molecules. Kluwer Academic Publishers, Netherlands, pp 3–20
Qadir M, Shams M (1997) Some agronomic and physiological aspects of salt tolerance in cotton (Gossypium hirsutum L.). J Agron Crop Sci 179:101–106
Quesada V, García-Martínez S, Piqueras P, Ponce MR, Micol JL (2002) Genetic architecture of NaCl tolerance in arabidopsis. Plant Physiol 130:951–963
Rieseberg LH, Archer MA, Wayne RK (1999) Transgressive segregation, adaptation and speciation. Hered 83:363–372
Rieseberg LH, Widmer A, Arntz AM, Burke JM (2003) The genetic architecture necessary for transgressive segregation is common in both natural and domesticated populations. Philos Trans R Soc Lond B 358:1141–1147
Saha S, Jenkins JN, Wu JX, McCarty JC, Gutierrez OA, Percy RG, Cantrell RG, Stelly DM (2006) Effects of chromosome-specific introgression in Upland cotton on fiber and agronomic traits. Genetics 172:1927–1938
SAS (2002) SAS systems for Windows 9.0., SAS Institute Inc., Cary, NC, USA
Shannon MC (1997) Adaptation of plants to salinity. Adv Agron 60:75–120
Tiwari RS, Picchioni G, Steiner RL, Hughs SE, Jones DC, Zhang JF (2013) Genetic variation in salt tolerance during seed germination in a backcross inbred line population and advanced breeding lines derived from Upland cotton × Pima cotton. Crop Sci (Accepted)
Turcotte EL, Percy RG, Feaster CF (1992) Registration of ‘Pima S-7’ American Pima cotton. Crop Sci 32:1291
UNESCO (2005) Did you know…? Facts and figures about water and salinization/desalination, UNESCO water e-newsletter no. 117: water and salinization/desalination. http://www.unesco.org/water/news/newsletter/117.shtml. Accessed 24 April 2013
Wang WX, Vinocur B, Altman A (2003) Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218:1–14
Yu JW, Yu SX, Zhang K, Fan SL, Song MZ, Zhai HH, Li XL, Huang SL, Zhang HW, Zhang JF (2012) Mapping quantitative trait loci for cottonseed oil, protein and gossypol content in a Gossypium hirsutum x Gossypium barbadense backcross inbred line population. Euphytica 187:191–201
Yu JW, Zhang K, Yu SX, Fan SL, Song MZ, Zhai HH, Li XL, Huang SL, Zhang HW, Li YH, Yang DG, Zhang JF (2013) Mapping quantitative trait loci for lint yield and fiber quality across environments in a Gossypium hirsutum x Gossypium barbadense backcross inbred line population. Theor Appl Genet 126:275–287
Zeng LH, Shannon MC, Lesch MC (2001) Timing of salinity stress affects rice growth and yield components. Agric Water Manag 48:191–206
Zhang LP, Lin GY, Foolad MR (2003) QTL comparison of salt tolerance during seed germination and vegetative growth in a Lycopersicon esculentum x L. pimpinellifolium RIL population. Acta Hort 618:59–67
Zhang JF, Sanogo S, Flynn R, Baral JB, Bajaj S, Hughs SE (2012) Germplasm evaluation and transfer of Verticillium wilt resistance from Pima (Gossypium barbadense) to Upland cotton (G. hirsutum). Euphytica 187:147–160
Zhang JF, Fang H, Zhou HP, Hughs SE, Jones DC (2013) Thrips resistance in cotton: germplasm evaluation, inheritance and QTL mapping. Proc. Beltwide Cotton Conf (in press)
Acknowledgments
The research was funded by USDA-ARS, Cotton Incorporated and New Mexico Agricultural Experiment Station. Brian Barrick provided assistance in the study.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Tiwari, R.S., Picchioni, G.A., Steiner, R.L. et al. Genetic variation in salt tolerance at the seedling stage in an interspecific backcross inbred line population of cultivated tetraploid cotton. Euphytica 194, 1–11 (2013). https://doi.org/10.1007/s10681-013-0927-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-013-0927-x