Abstract
Nowadays, heart diseases are significantly contributing to deaths all over the world. Thus, heart-disease prediction has garnered considerable attention in the medical domain globally. Accordingly, machine-learning algorithms for the early prediction of heart diseases were developed in several studies to help physicians design medical procedures. In this study, a hybrid genetic algorithm (GA) and particle swarm optimization (PSO) optimized approach based on random forest (RF), called GAPSO-RF, is developed and used to select the optimal features that can increase the accuracy of heart-disease prediction. The proposed GAPSO-RF implements multivariate statistical analysis in the first step to select the most significant features used in the initial population. After that, a discriminate mutation strategy is implemented in GA. GAPSO-RF combines a modified GA for global search and a PSO for local search. Moreover, PSO achieved the concept of rehabbing individuals that had been refused in the selection process. The performance of the proposed GAPSO-RF approach is validated via evaluation metrics, namely, accuracy, specificity, sensitivity, and area under the receiver operating characteristic (ROC) curve by using two datasets from the University of California, namely, Cleveland and Statlog. The experimental results confirm that the GAPSO-RF approach attained the high heart-disease-prediction accuracies of 95.6% and 91.4% on the Cleveland and Statlog datasets, respectively. Furthermore, the proposed approach outperformed other state-of-the-art prediction methods.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The heart pumps blood to the entire human body. Coronary arteries are the blood-vessels that transport oxygenated blood to the heart [36]. The shrinking of coronary arteries is the primary cause of heart failure (HF). Heart diseases are one of the major reasons of human mortality, as reported by the World Health Organization. In 2013, heart diseases caused the highest number of deaths globally, at approximately 17.3 million. Similarly, in 2016, approximately 17.6 million deaths were attributed to heart diseases, amounting to a rise of 14.5% from 2006 [10]. Moreover, patients with HF suffer from other symptoms as well, including difficulty in breathing, weakness, and swollen feet [14]. Heart diseases may be managed or controlled if trained medical professionals detect them at their early stages, thereby enabling them to make the correct decision. Therefore, early detection of heart diseases is critical to improving HF symptoms and extending the lives of patients [8]. The medical history of a patient includes a substantial number of features. However, not all these features may be equally significant, and some may even be redundant. Additionally, using all the features at once deteriorates the performance of diagnosis. Most research-based on heart-disease prediction focused on two factors: selecting the best features while dismissing the irrelevant ones and choosing an appropriate classifier. Therefore, the prediction methods are aimed at selecting the optimal features and an appropriate classifier. Recently, machine-learning-based methods have improved the quality of our lives, especially in the medical domain [2, 5, 7, 20, 46, 48, 49, 53].
Many research papers have used machine learning to diagnose heart disease and predict whether a patient has heart disease [1, 25, 28,29,30, 51]. Recently, Amin et al. [5] presented a hybrid technique that comprised Naïve Bayes (NB), logistic regression, and feature selection (FS). Revett et al. [44] deployed the use of rough sets to determine the information content of each subset of the feature space. Furthermore, support vector machines (SVMs) were applied in some researches including [48, 49]. Saqlain et al. [46] implemented Fisher score for FS and SVM for classification. Saifudin et al. [45] applied bagging based on random forest (RF) to improve classification accuracy of heart disease. Subsequently, Gupta et al. [19] used Yule-Walker (YW) and Principal Component Analysis (PCA) for R-peak Detection in Electrocardiogram (ECG) signal, during the detection process regular and abnormal signals were considered. The results obtained using PCA with YW carried out the results using PCA without YW. Besides, FS was also implemented in other domains [27] to increase the classification accuracy by presenting a multi-layer hybrid technique to detect peer to peer botnets. A decision tree algorithm is applied for feature selection to extract the most relevant features and ignore the irrelevant features. They achieved high accuracy by using a decision tree algorithm and their experiments prove the benefits of using multi-layer instead of single layer. In addition, Reddy et al. [41] proposed an approach for diabetes diagnosis, the authors used Locality Preserving Projection (LPP) algorithm for feature reduction and Firefly-BAT (FFBAT) optimization algorithm with artificial neural network (ANN) for diabetes disease classification. The results have proved that the proposed classification framework outperforms the existing method by achieving better accuracy. In conclusion, FS is the most crucial step in increasing the accuracy of heart-disease diagnosis. For example, a doctor might decide regarding a patient who suffers from HF based on classification implemented using the selected features. The previous researches gave more attention to improving and developing classification methods than selecting the best features. In addition, it needs to improve the accuracy rate.
The objectives of this work are: 1) Select the best features, 2) Improve the heard disease prediction accuracy, and 3) Improve the complexity time. Therefore, we introduce an efficient, hybrid genetic algorithm (GA) and particle swarm optimization (PSO) approach based on random forest (RF) for optimizing the FS process to select the crucial features that increase the accuracy of heart-disease diagnosis. The main contribution of this paper is to develop a hybrid approach, called GAPSO-RF, for heart-disease prediction. First, a discriminate mutation strategy based statistical analysis is applied to be used in the adaptive mutation operator for GA. Second, a modified genetic algorithm with PSO supported by the RF algorithm is used to select the best features. PSO is used to target the rejected individuals of each generation to fulfill the concept of rehabbing the rejected individuals, maximizing the utilization of all individuals in each generation. Finally, the proposed GAPSO-RF is validated via evaluation metrics, namely, accuracy, specificity, and area under the receiver operating characteristic (ROC) curve by using two heart-disease datasets from the University of California (UCI), Irvine, machine learning repository [13], namely, Cleveland and Statlog. Experimental findings suggest that the proposed GAPSO-RF achieves high prediction accuracies.
The rest of this paper is structured as follows. Section 2 illustrates the related work. The materials and proposed approach are discussed in Section 3, including the description of both the datasets, background concepts related to FS, and classification process. The experimental results are provided in Section 4, including a comparative analysis of our method with those in the literature. Finally, the conclusions are drawn in Section 5.
2 Related works
Recent researches have been focused on FS, prediction, and increasing the heart-disease-prediction accuracy. This section overviews the recently published related researches. Lately, Mohammad S. Amin et al. [5] developed a heart-disease-prediction model by using the identified best features and data-mining algorithms on the Cleveland dataset. Subsequently, Saqlain et al. [46] employed Fisher score and the Matthews correlation coefficient as an FS algorithm and SVM for binary classification to diagnose heart diseases on several datasets. Purnomo et al. [39] applied FS in the form of backward elimination on NB to increase classification accuracy on heart-disease from 84.29% to 89.45%. Besides, a fuzzy algorithm was used as another solution by Vivekanandan and lyengar [51]. Priyatharshini and Chitrakala [38] developed a self-learning fuzzy rule-based system to predict heart disease, the authors achieved an overall accuracy 90.7%. Subsequently, Halder er al. [21] implemented computerized diagnosis system using Rough set classifier from multi-lead ECG signal for the classification of myocardial infarction (MI) disease. Dwivedi [15] applied different algorithms, namely, ANN, SVM, logistic regression, k-nearest neighbors (KNN), classification tree, NB, and achieved the highest accuracy in logistic regression. Recently, Krishnaiah et al. [29] proposed a fuzzy KNN approach by presenting an exponential membership function with standard deviation, and they calculated the mean of the attributes measured. Buettner and Schunter [11] performed classification using the RF algorithm, which they validated on the Cleveland dataset. Notably, their method was not an FS approach. However, several studies, including [35, 50], used GAs for performing FS. Ismaeel et al. [22] proposed an improved extreme learning machine algorithm and implemented it on the Cleveland dataset; their algorithm performed better than back-propagation neural networks. El-Bialy et al. [16] performed FS using fast decision tree and C4.5 pruning tree algorithms. Saxena et al. [47] used decision-trees for rule generation. Reddy et al. [43] implemented an adaptive genetic algorithm with fuzzy logic to predict heart disease based on a rough set for features selection. However, the previous studies on heart-disease prediction still lack optimizing FS and using an appropriate classifier to enhance the performance of the heart-disease classification. Table 1 provides a summary of the related methods included in this study.
However, the previous studies on heart-disease prediction still lack optimizing FS and using an appropriate classifier to enhance the performance of the heart-disease classification. Although several studies proposed different FS algorithms, they did not considerably focus on GA. While GA is best known for searching and finding the best subset features from the original features that enhance the classification. In addition, FS is the most important factor in improving the accuracy of heart-disease diagnosis to help doctors make the correct decision. Therefore, we aim to select the best features by using the GAPSO-RF approach. Which selects the best features using GA and PSO and at the same time enhance the performance of the heart-disease classification with RF algorithm.
3 Materials and the proposed approach
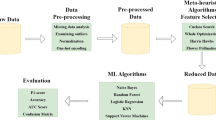
We aim to select the best features to increase the heart-disease-diagnosis accuracy. Thus, an GAPSO-RF based FS approach is proposed. Before the FS process, we implement discriminate mutation strategy based statistical analysis to be used in adaptive mutation operator in GA. After that, the features ranges are normalized by applying min--max normalization. During the FS process, the proposed GAPSO-RF utilizes GA to search for a set of optimal features by optimizing the hyper-parameters of the GA and the modified selection operator. The rejected individuals from selection are passed to the PSO for reformation. The population of PSO will be made of these rejected individuals who will connect to update their position and velocity to achieve the best possible result from the non-fit individuals. The best individuals of PSO will be injected into the new population of GA. The fitness function in both GA and PSO is optimized using an optimized RF classifier to increase the classification accuracy. The overall workflow of the proposed approach is illustrated in Fig. 1. Four tasks have to be performed for prediction: (1) statistical analysis and data pre-processing, (2) GAPSO-RF utilization, (3) RF-based classification, and (4) performance measurement. In the following subsections, we describe the datasets, and then each step of the proposed approach is discussed.
3.1 Datasets description
In the proposed approach, two datasets, namely, Cleveland and Statlog, from the UCI machine-learning repository are used [13]. Table 2 lists the features of both the datasets. The (Num) variable represents two values of the heart-disease diagnosis: 0 means healthy (the patient has no heart disease), and 1 means unhealthy (the patient has a heart disease). As shown in Fig. 2, in the Cleveland dataset, 165 records have the value of (1), and 138 have the value of (0). In addition, in the Statlog dataset, 120 records have the value of (1), and 150 have the value of (0).
3.2 Statistical analysis and pre-processing
3.2.1 Multivariate statistic analysis
The first step is to analyze the conditional mean and variance for each attribute conditioning on ‘Num = 1’ and ‘Num = 0’, and calculate the T2 metric for each attribute, where T2 metric is defined as follows:
where \( {\overline{X}}_1 \) and \( {\overline{X}}_0 \) are the mean for Num equals 1 and 0, respectively, and Sp is defined as follows:
where n1 and n0 are the sample numbers when Num equals 1 and 0, respectively. In addition, S1 and S0 are the standard deviations for Num equals 1 and 0, respectively. Tables 3 and 4 report the statistical analysis for all the attributes in the selected datasets when Num equals 1 and 0, respectively. Moreover, the T2 metric is calculated in Tables 5 and 6 for Cleveland and Statlog datasets, respectively.
As depicted in Table 5, the attributes (Age, Trestbps, Chol, Thalach, and Oldpeak) of Cleveland dataset have the most considerable variance. It is because these attributes are continues, and a considerable variance is expected for continuous attributes. The attributes (Sex, Fbs, and Restecg) are non-continuous, and have the slightest variances, so they provide much less information according to the entropy theory. The remaining attributes (Cp, Exang, Slope, Ca, and Thal) are non-continuous, and have the highest variances, providing the essential information for classification. Further correlation analysis is implemented for (Cp, Exang, Slope, Ca, and Thal) and shows some correlation between (exang) and the other four attributes as shown in Table 7. After excluding (Exang), the remaining four attributes (Cp, Slope, Ca, and Thal) are the most significant. These four attributes will have a lower mutation probability (e.g., 10−3) throughout GA evolution.
Table 6 presents the T2 metric for each attribute in the Statlog dataset. The attributes (Thalach, Chol, Thal, Trestbps, and Age) have the most considerable variance. The attributes (Sex, Fbs, and Restecg) are non-continuous, and have the slightest variances, so they provide much less information according to the entropy theory. The remaining attributes (Cp, Exang, Oldpeak, Slope, and Ca) are non-continuous, and have the highest variances, providing the essential information for classification. Further correlation analysis for (Cp, Exang, Oldpeak, Slope, and Ca) shows some correlation between (Exang) and the other four attributes as shown in Table 8. After excluding (Exang), the remaining four attributes (Cp, Oldpeak, Slope, and Ca) are the most significant. These four attributes will have a lower mutation probability (e.g., 10−3) throughout GA evolution.
3.2.2 Discriminate mutation strategy in genetic algorithm
In the current GA algorithm, the individual dimension is 13 (as there are 13 attributes), so it requires a population size of 213 = 8192 individuals to cover all possible combinations. An improvement is always selecting the attributes with the most critical information to be endowed with less mutation probability (i.e., 10−3). In comparison, the remaining attributes will have a higher mutation probability to explore more individuals with higher fitness. In the simulation, the sets (Cp, Slope, Ca, and Thal) and (Cp, Oldpeak, Slope, and Ca) are the most significant attributes for Celeveland and Statlog, respectively, which have less mutation probability, (i.e., 10−3). The remaining attributes will have higher but equal mutation probabilities. The initialization of the population at the start of the GA should also be modified using this discriminate mutation strategy. In the initial population, the individual should always have the most significant attributes.
3.2.3 Data pre-processing
The data are normalized before performing FS. To that end, we normalize the dataset values. This process has been gaining importance because all features may have different data types, and it eliminates the numerical difficulties due to the different range of values during the computation process. In the proposed approach, we implemented min--max normalization, a technique that converts a value a to \( \overset{`}{a} \) in the range of [max _ new − min _ new] as follows:
where from min _ new to max _ new denotes the range of the transformed values. We implemented min _ new = 0 and max _ new = 1. Subsequently, these transformed values were used as input for the FS method.
3.3 Hybrid modified genetic algorithm and particle swarm optimization
FS, which is a method of selecting reduced number of appropriate features, enhances the classification by determining the best subset of features from the set of original features. It eliminates unnecessary features, thereby lowering the computational and memory costs. It involves selecting a subset of features t from the total features T based on a particular optimization criterion. GA combined with PSO was used as an FS approach to search for optimal solutions. GA was introduced by John Holland in 1975. Although they can be used for solving both search and optimization problems, they are best known for solving the latter (Holland, 1992). A GA is a search heuristic that mimics the natural evolution process. It is regularly used to produce useful solutions for the problems related to search and optimization. Notably, GA belongs to the broader category of evolutionary algorithms (EAs). In several real-world optimization projects, EAs have proved to be the most efficient solution. Substantially, Holland supposed that the population size is limitless, that the fitness function correctly represents the convenience of a solution, and that the correlations between the genes are very small, which leads to problems [9]. Population size is limited, impacting the GA’s sampling capacity and efficiency.
PSO were introduced by Eberhart and Kennedy [26] in 1995. It is a population-based optimization technique that is inspired by the behavior of fish schooling or bird flocking. PSO algorithm is one of several types of swarm intelligence algorithms. One of several PSO’s main advantages is that it is computationally inexpensive due to its low system requirements [37]. Using a local search approach in combination with GA solve much of the hindrances that occur due to the finite population size. Hybridization has proven to be an efficient way to construct capable genetic algorithms. By adding new genes, a local search approach with GA helps neutralize much of the challenges that exist because of the limited population size as well as the genetic drift dilemma [6]. A GA uses the laws of genetics as its paradigm for implementing problem-solving on a population (P) of individuals. Each individual is characterized by a set of variables called genes. To build a chromosome, genes are combined into a string. Therefore, each solution is represented by a chromosome.
Chromosomes Ck, where k = (1, …, P), are encoded in a binary vector Bk of length n. Binary encoding is used to identify whether a feature is selected for input or not. The group of all the chromosomes is referred to as population. In the initial population, the individual should always have the four most significant attributes, (Cp, Slope, Ca, and Thal). After that, a GA accomplishes its task via four basic operations: modified selection, crossover, modified mutation, and fitness calculation. We believe that (non-fit individuals) can contain good genes that can direct the search process’ cursor to locations in the search space where significant improvements can be found. Accordingly, in the selection operation, the rejected chromosomes (non-fit individuals) are passed to PSO for reformation as GA searches for good chromosomes, not good genes, and the best individuals (fittest individuals) are selected based on the value of the fitness function, which is calculated in both GA and PSO using the RF algorithm and with high efficiency to survive to next generation. Moreover, RF prevents over-fitting, which is one of the main challenges in heart-disease prediction. Therefore, in the proposed GAPSO-RF, the RF classifier is used with GA to select the best features. Algorithm 1 presents the hybrid approach using modified genetic algorithm and PSO. RFs comprise many individual decision-trees, which function as an ensemble. An algorithm that can construct many small decision-trees using a few features is considered computationally cheap. If we can create several small, weak decision-trees in parallel, then by averaging or taking the majority vote, we can combine the trees to form a single, strong learner. Practically, RFs are the most effective learning algorithms to date. The RF algorithm is illustrated in Algorithm 2. In the following subsections, we detail the GAPSO-RF processes, namely, selection, crossover, and mutation.


3.3.1 Modified tournament-selection operator
In a GA cycle, the initial population is set to be 50 and the maximum iteration number to be 30 generations. Subsequently, we begin the tournament-selection process, which is critical to selecting the best individuals, which have been appraised for their fitness value, from the current generation for reproduction or to be survived in the successive generation and the rejected individuals are passed to the PSO for reformation. The population of PSO will be made of these rejected individuals who will connect with one another to update their position and velocity to achieve the best possible result from the non-fit individuals. The best individuals of PSO will be injected to the new population of GA.
The fitness function was applied in GA by implementing the RF classifier, evaluating the score of each solution, and observing how close the solution was to the one we needed. To calculate the fitness function, a chromosome must first be decoded in the binary representation. Tournament selection was applied with a size of 0.26. Although tournament selection is equivalent to rank selection with respect to the selection pressure, it is more effective in computation and more appropriate for parallel implementation [33].
The fitness of the individuals is calculated by 1) transforming the feature space of the dataset, and 2) applying k-fold cross-validation or holdout validation and obtaining a high accuracy score from the RF classifier. The selection probability of each individual is calculated as follows:
where Ps(c) and v(c) denote the probability of selection and the fitness value for the cth chromosome, respectively.
3.3.2 Crossover operator
In this process, two parent chromosomes are used to construct a new chromosome on the basis of crossover probability, which is 0.5 in the experiments. The constructed chromosome has a better string than those of its parent chromosomes. The following are the steps of the crossover:
-
1)
A combination of two individual strings is chosen with the assistance of the reproduction operator.
-
2)
A cross-site is randomly picked alongside the length of the string.
-
3)
swapping the positions of values between the two strings.
3.3.3 Modified mutation operator
Upon the completion of the crossover process, the strings undergo mutation, which is the random change in the value of a gene. Mutation means that we flip a single bit from 0 to 1 or vice versa. The mutation operator is used to obtain a better solution by changing the current one. Mutation prevents the GA from being stuck in a local minimum. The mutation operator is modified by implementing the discriminate mutation strategy based statistical analysis as illustrated in Section 3.2.2.
3.4 Random forest classification
In the proposed approach, the RF algorithm is used for binary classification. RF constructs many decision-trees during the training period and generates a class that has a mean prediction. The hyper-parameters of RF were tuned using a grid search. A wide range of parameter values were implemented in grid search as shown in Table 9. The best set of parameters extracted from the grid search was used to train random forest to get max classification accuracy.
We specify the number of random trees as 1000, maximal depth as 10 using confidence 0.5 in vote strategy and Gini impurity in criterion (split criterion); pruning and pre-pruning are applied; minimal leaf size is 2; the minimal size for splitting is 4. The Gini impurity is calculated as follows:
where C denotes the number of classes and p(j) the probability of choosing a class j data point.
3.5 Performance measures
Four measures were implemented to assess the performance of the classification models: accuracy, recall, precision, receiver operating characteristic (ROC), and area under the ROC curve (AUC). Accuracy represents the rate of correctness of a classifier. Therefore, we take the sum of true positive (TP) records and true negative (TN) records and then divide by the total number of records which represents the sum of TN, TP, false negative (FN) and false positive (FP); thus, accuracy denotes the ratio of the number of correctly predicted records to the total number of records, as shown in Eq. (6). Recall represents the rate of values that measures positive records that the classifier correctly predicted. Moreover, it is called true positive rate (TPR) or sensitivity. Thus, recall is calculated as shown in Eq. (7). Precision is the ratio of TP records to the total of positive predicted records, as shown in Eq. (8). The ROC curve is a graph of TPR versus false positive rate (FPR), where TPR is on the y-axis and FPR on the x-axis. The AUC metric is used to calculate AUC, and it describes the separability measurement or degree. It informs how the model can identify among classes.
4 Experiments results and discussion
In this section, two public datasets, namely, Cleveland and Statlog, are used to evaluate the proposed approach, and then the classification performance of our approach is compared with those of other state-of-the-art methods. Moreover, the proposed approach will be compared with the methods that implement GA and with those that do not implement GA. In addition, we will discuss the complexity of the proposed approach.
4.1 Experimental setup
In this section, two types of experiments are implemented on the Cleveland and Statlog datasets to assess the efficacy of the proposed model. All the computations are performed on Google CoLab, which provides GPU Tesla k80 with 12 GB of GDDR5 VRAM, and Intel Xeon Processor with two 2.20-GHz cores and 13 GB RAM. Moreover, the Python programming software package scikit-learn is used for the experiments.
4.2 Results of the Cleveland dataset
The model was applied on the Cleveland heart-disease dataset, which had 13 features. All the 303 heart-disease records of the dataset were considered. To assess the classification performance, the results obtained from the experiments of the proposed model were compared with those of other state-of-the-art methods in terms of heart-disease prediction. In the first experiment, throughout cross-validation, the data records were divided into 10 folds; one-fold was used in testing, and the remaining nine folds were used in training. The data were divided into folds via stratified sampling, meaning that the class distribution (defined by the label attribute) in the subsets/folds was the same as that in the complete dataset. Finally, the result was obtained by averaging all the 10 iterations.
In the second experiment, we performed the train/test holdout validation. The data were split as 70% for training and 30% for testing. The model was trained on 212 records and tested on the remaining 91 as unseen data. The primary reason behind using this distribution is to satisfactorily compare our approach with those in other researches on the same dataset. We ran the same experimental procedure five times, following which the mean of the five results was calculated. Table 10 compares the results of these two experiments with those of recent researches. Evidently, the proposed approach achieves better classification results than those of most methods. The experimental results on the Cleveland dataset confirm that the proposed approach achieves the accuracy rates of 87.8% and 95.6% for 10-fold and holdout (TR = 70%) validations, respectively. In the 10-fold validation, the proposed approach increases the average accuracy by 6.61%, 5.62%, and 2.67% over Saqlain et al. [46], Shah et al. [48], and Mathan et al. [32], respectively. The results of the other experiment (holdout) demonstrate that the proposed increases the average accuracy by 7.26%, 3.4%, and 2.27% over Gokulnath and Shantharajah [18], Ali et al. [3], and Ali et al. [4], respectively. Figure 3a and b depict the ROC analysis for the 10-fold and holdout (TR = 70%) validations, respectively. Table 11 compares results of using subset features selected of GA for Cleveland dataset with other models. The results show that RF overcomes the other models.
4.3 Results of the Statlog dataset
We compare our approach with several benchmark approaches on the Statlog heart-disease dataset, which contained 13 features. All the 270 heart-disease records of the dataset were considered. We performed the same experiments on the Statlog dataset as those performed on the first dataset, i.e., Cleveland. First, the data records were partitioned into 10 folds, and the results were obtained by calculating the mean of all the ten iterations. Second, the data were split as 70% for training and 30% for testing (i.e., holdout (TR = 70%). The model was trained on 189 records and tested on the remaining 81 records as unseen data. The primary reason behind using this distribution was to satisfactorily compare our approach with those in other researches on the same dataset. We performed the same experimental procedure five times and recorded the average of the five results.
Table 12 compares the results of the proposed approach with those of the recent state-of-the-art heart-disease-prediction methods. Evidently, our approach obtains the accuracy rates of 87.78% and 91.4% for the 10-fold and holdout (TR = 70%) validations, respectively, the best results achieved on the Statlog dataset thus far. In the 10-fold validation, the proposed approach increases the average accuracy by 11.18% and 3.78% over El-Bialy et al. [16] and Rado et al. [40], respectively. In the other experiment (holdout (TR = 70%)), the results proved that the proposed approach increases the average accuracy by 12.62% and 1.4% over Long et al. [30] and Karthikeyan and Kanimozhi [24], respectively.
Figure 4a and b depict the ROC analysis for the 10-fold and holdout (TR = 70%) validations, respectively. Table 13 compares results of using subset features selected of GA for statlog dataset with other models. The results show that RF overcomes the other models.
Although several studies have implemented FS in their proposed methods, little attention has been given to optimizing fitness function in GA. In our proposed approach, we used RF as a fitness function in GA to get the maximum value of classification accuracy. We tuned the hyper-parameters of RF using grid search; after that, we applied pruning and pre-pruning. Besides that, we implemented the experiments in GA different hyper-parameters and different types of crossover, mutation and selection as shown in Fig. 5. In the crossover, we applied uniform as it achieved better results than one point. Moreover, we used tournament selection as it produced better results than roulette wheel.
Different hyper-parameters which affect the efficiency of GA. Red and green curves represent the max and average fitness, respectively. a number of generations = 50, population size = 50, crossover rate = 0.5 and mutation rate = 0.07. b number of generations = 50, population size = 50, crossover rate = 0.5 and mutation rate = −0.4. c number of generations = 30, population size = 50, crossover rate = 0.5 and mutation rate = 0.07. d number of generations = 30, population size = 50, crossover rate = 0.5 and mutation rate = 0.08
Table 14 summarizes the performance-evaluation results for both 10-fold and holdout (TR = 70%) validations on the Statlog dataset. As seen from Table 14, our proposed approach with the optimal selected features achieved a better performance than that achieved upon using all the features at once. Additionally, our proposed approach increases the average accuracy on the Statlog dataset by 5.93% and 7.45% in the 10-fold and holdout (TR = 70%) validations, respectively.
4.4 Effectiveness of FS
In this subsection, the performance of the FS process in GA is evaluated. FS improves the performance of the proposed approach compared with using all the features at once. The features sets of the Cleveland and Statlog datasets are reduced by 46.15% and 30.77%, respectively. From Table 15, it is evident the proposed approach decreased the number of features. The features selected in Cleveland dataset are 7 features (Cp, Fbs, Restecg, Exang, Slope, Ca, and Thal) and for statlog dataset the features selected are 9 features (Age, Sex, Cp, Fbs, Thalach, Exang, Slope, Ca, and Thal). The overall measurement results for the Cleveland dataset both with and without FS on GA are summarized in Table 16. As previously mentioned, the experiment was performed twice (10-fold and holdout (TR = 70%)). From the experimental results in Table 16, one can see that the proposed approach with selected optimal features achieves better performance than that achieved upon using all the features at once. Our proposed approach increases the average accuracy on the Cleveland dataset by 4.33% and 6.59% in the 10-fold and holdout (TR = 70%) validations, respectively.
4.5 Time complexity
In this subsection, we compare the time complexity of our proposed approach GAPSO-RF with GA-based different models. Table 17 shows the comparison of computational cost among the approaches on the Cleveland dataset. This table records the complexity time (i.e., FS and classification), number of generations, and prediction accuracy. We can see that the complexity time of our approach GAPSO-RF is not the best. However, as concluded from Table 17, our proposed approach achieves the best prediction accuracy compared to other GA-based methods. In addition, the proposed approach reached the best rate of 87.80% by the minimum number of generations 30.
First, the proposed GAPSO-RF approach improves the conventional GA as follows: 1) Reduce the number of generations by 70%, 2) Execution time improved by 65.87%, and 3) Improve the prediction accuracy by 0.67%. Second, a convolutional neural network (CNN) is good in feature selection, as mentioned in [17, 52]. However, it consumes a higher computation cost. Results show that GA-based CNN has a higher computational cost than others. In addition, the proposed GAPSO-RF outperforms GA-CNN in prediction accuracy and execution time. In the future, the CNN model can be used for different types of datasets (e.g., electrocardiogram (ECG) signals and images). The implementation of a discriminate mutation strategy in GA-based statistical analysis and the implementation of the PSO in local search are reasons to reduce the number of generations and thus reduce the execution time. Despite this, the execution time in our approach is relatively large. In the future, we intend to develop an efficient feature selection method with low complexity and high performance. Third, GA-NB achieves the best execution time due to the number of iterations in PSO and RF. Nevertheless, we outperform this approach by 5.88% in the average prediction accuracy.
5 Conclusion
We presented a GAPSO-RF-based FS approach with an RF classifier as the base of a fitness function to select significant features to increase the accuracy of heart-disease diagnosis. The proposed approach achieved high accuracy of 95.6% and 91.4% on the Cleveland and Statlog datasets, respectively. After that, the results of the proposed FS method are compared with the results without using FS and found that it outperforms in accuracy. Moreover, it outperformed the existing state-of-the-art methods on the same datasets. Furthermore, a comparative analysis is performed between GAPSO-RF and conventional GA and found that our proposed approach outperformed conventional GA. Additionally, we protected our model from overfitting by using the RF algorithm for classification. Hence, our experimental results confirmed that the proposed approach enhanced the decision-making process of the practitioners during heart-disease diagnosis.
The following are some of the limitations of this research: First, more classifiers should be evaluated to have a more extensive evaluation of the results. Second, the proposed model’s key drawbacks are its high computational cost and temporal complexity, as it is based on the wrapper feature selection strategy. More studies need to be conducted to address the limitations of the proposed approach in the future. First, multi-objective genetic algorithm can be applied. Second, to overcome small-data limitations in heart-disease prediction, we plan to use in future work surrogate data or merging different heart-disease datasets. Finally, for electrocardiogram (ECG) signals and images, further study can be carried out for improving the features selection by using convolutional neural network (CNN). Moreover, we intend to develop an efficient classifier to improve the performance.
References
Abdel-Basset M, Gamal A, Manogaran G, Long HV (2019) A novel group decision making model based on neutrosophic sets for heart disease diagnosis. Multimed Tools Appl:1–26
Adler ED, Voors AA, Klein L, Macheret F, Braun OO, Urey MA, Zhu W, Sama I, Tadel M, Campagnari C, Greenberg B, Yagil A (2020) Improving risk prediction in heart failure using machine learning. Eur J Heart Fail 22:139–147
Ali L, Niamat A, Khan JA, Golilarz NA, Xingzhong X, Noor A, Nour R, Bukhari SAC (2019) An optimized stacked support vector machines based expert system for the effective prediction of heart failure. IEEE Access 7:54007–54014
Ali L, Rahman A, Khan A, Zhou M, Javeed A, Khan JA (2019) An automated diagnostic system for heart disease prediction based on x2 statistical model and optimally configured deep neural network. IEEE Access 7:34938–34945
Amin MS, Chiam YK, Varathan KD (2019) Identification of significant features and data mining techniques in predicting heart disease. Telematics Inform 36:82–93
Asoh H, Mühlenbein H (1994) On the mean convergence time of evolutionary algorithms without selection and mutation. In: International conference on parallel problem solving from nature, pp 88–97
Atal DK, Singh M (2020) A dictionary matrix generation based compression and bitwise embedding mechanisms for ECG signal classification. Multimed Tools Appl 79:13139–13159
Banerjee D, Thompson C, Kell C, Shetty R, Vetteth Y, Grossman H et al (2017) An informatics-based approach to reducing heart failure all-cause readmissions: the Stanford heart failure dashboard. J Am Med Inf Assoc 24:550–555
Beasley D, Bull DR, Martin RR (1993) An overview of genetic algorithms: part 1, fundamentals. Univ Comput 15:56–69
Benjamin EJ, Muntner P, Bittencourt MS (2019) Heart disease and stroke statistics-2019 update: a report from the American Heart Association. Circulation 139:e56–e528
Buettner R, Schunter M (2019) Efficient machine learning based detection of heart disease, presented at the IEEE international conference on E-health networking, Application & Services (HealthCom), Bogota, Colombia, Colombia
Chitra R, Seenivasagam V (2015) Heart disease prediction system using intelligent network. In: Power electronics and renewable energy systems. Springer, pp 1377–1384
Dua D, Graff C (2017) UCI Machine Learning Repository. Available: http://archive.ics.uci.edu/ml
Durairaj M, Sivagowry S (2014) A pragmatic approach of preprocessing the data set for heart disease prediction. Int J Innov Res Comput Commun Eng 2:6457–6465
Dwivedi AK (2018) Performance evaluation of different machine learning techniques for prediction of heart disease. Neural Comput Appl 29:685–693
El-Bialy R, Salamay MA, Karam OH, Khalifa ME (2015) Feature analysis of coronary artery heart disease data sets. Procedia Comput Sci 65:459–468
Garcia-Garcia A, Orts-Escolano S, Oprea S, Villena-Martinez V, Garcia-Rodriguez J (2017) A review on deep learning techniques applied to semantic segmentation. arXiv preprint arXiv:1704.06857
Gokulnath CB, Shantharajah SP (2018) An optimized feature selection based on genetic approach and support vector machine for heart disease. Clust Comput 22:1–11
Gupta V, Mittal M (2019) R-peak detection in ECG signal using yule–Walker and principal component analysis. IETE J Res 67:1–14
Gupta V, Mittal M, Mittal V (2020) Performance evaluation of various pre-processing techniques for R-peak detection in ECG signal. IETE J Res:1–16
Halder B, Mitra S, Mitra M (2019) Classification of complete myocardial infarction using rule-based rough set method and rough set explorer system. IETE J Res:1–11
Ismaeel S, Miri A, Chourishi D (2015) Using the extreme learning machine (ELM) technique for heart disease diagnosis. In: IEEE Canada international humanitarian technology conference (IHTC2015), Ottawa, ON, Canada, pp 1–3
Jha SK, Pan Z, Elahi E, Patel N (2019) A comprehensive search for expert classification methods in disease diagnosis and prediction. Expert Syst 36:e12343–e12343
Karthikeyan T, Kanimozhi V (2017) Deep learning approach for prediction of heart disease using data mining classification algorithm deep belief network. Int J Adv Res Sci Eng Technol 4:3194–3201
Kaur P, Kumar R, Kumar M (2019) A healthcare monitoring system using random forest and internet of things (IoT). Multimed Tools Appl 78:19905–19916
Kennedy J, Eberhart R (1995) Particle swarm optimization. In: Proceedings of ICNN'95-international conference on neural networks, pp 1942–1948
Khan RU, Zhang X, Kumar R, Sharif A, Golilarz NA, Alazab M (2019) An adaptive multi-layer botnet detection technique using machine learning classifiers. Appl Sci 9:2375
Kohli R, Garg A, Phutela S, Kumar Y, Jain S (2021) An improvised model for securing cloud-based E-Healthcare systems. In: IoT in Healthcare and ambient assisted living. Springer, pp 293–310
Krishnaiah V, Narsimha G, Chandra NS (2015) Heart disease prediction system using data mining technique by fuzzy K-NN approach, vol 337. Springer, Cham
Long NC, Meesad P, Unger H (2015) A highly accurate firefly based algorithm for heart disease prediction. Expert Syst Appl 42:8221–8231
Luo M, Wu K (2020) Heart rate prediction model based on neural network. IOP Conf Ser Mater Sci Eng 715:012060–012060
Mathan K, Kumar PM, Panchatcharam P, Manogaran G, Varadharajan R (2018) A novel Gini index decision tree data mining method with neural network classifiers for prediction of heart disease. Des Autom Embed Syst 22:225–242
Mitchell M (1998) An introduction to genetic algorithms. MIT press
Mukherjee S, Kapoor S, Banerjee P (2017) Diagnosis and identification of risk factors for heart disease patients using generalized additive model and data mining techniques. J Cardiovasc Dis Res 8:137–144
Paul AK, Shill PC, Rabin MRI, Akhand MAH (2016) Genetic algorithm based fuzzy decision support system for the diagnosis of heart disease. In: 5th international conference on informatics, electronics and vision (ICIEV), Dhaka, Bangladesh, pp 145–150
Polat K, Şahan S, Güneş S (2007) Automatic detection of heart disease using an artificial immune recognition system (AIRS) with fuzzy resource allocation mechanism and k-nn (nearest neighbour) based weighting preprocessing. Expert Syst Appl 32:625–631
Prado R, García-Galán S, Yuste AJ, Expósito JM (2010) A fuzzy rule-based meta-scheduler with evolutionary learning for grid computing. Eng Appl Artif Intell 23:1072–1082
Priyatharshini R, Chitrakala S (2019) A self-learning fuzzy rule-based system for risk-level assessment of coronary heart disease. IETE J Res 65:288–297
Purnomo A, Barata MA, Soeleman MA, Alzami F (2020) Adding feature selection on Naïve Bayes to increase accuracy on classification heart attack disease. J Phys Conf Ser 1511:012001–012001
Rado O, Ali N, Sani HM, Idris A, Neagu D (2019) Performance analysis of feature selection methods for classification of healthcare datasets. In: Intelligent computing-proceedings of the computing conference, pp 929–938
Reddy GT, Khare N (2017) Hybrid firefly-bat optimized fuzzy artificial neural network based classifier for diabetes diagnosis. Int J Intell Eng Syst 10:18–27
Reddy GT, Khare N (2017) An efficient system for heart disease prediction using hybrid OFBAT with rule-based fuzzy logic model. J Circ Syst Comput 26:1750061
Reddy GT, Reddy MPK, Lakshmanna K, Rajput DS, Kaluri R, Srivastava G (2020) Hybrid genetic algorithm and a fuzzy logic classifier for heart disease diagnosis. Evol Intel 13:185–196
Revett K, Gorunescu F, Salem A-B, El-Dahshan E-S (2009) Evaluation of the feature space of an erythematosquamous dataset using rough sets. Ann Univ Craiova-Math Comput Sci Ser 36:123–130
Saifudin A, Nabillah UU, Yulianti, Desyani T (2020) Bagging technique to reduce misclassification in coronary heart disease prediction based on random forest. J Phys Conf Ser 1477:032009–032009
Saqlain SM, Sher M, Shah FA, Khan I, Ashraf MU, Awais M, Ghani A (2019) Fisher score and Matthews correlation coefficient-based feature subset selection for heart disease diagnosis using support vector machines. Knowl Inf Syst 58:139–167
Saxena K, Sharma R, others (2016) Efficient heart disease prediction system. Procedia Comput Sci 85:962–969
Shah SMS, Batool S, Khan I, Ashraf MU, Abbas SH, Hussain SA (2017) Feature extraction through parallel probabilistic principal component analysis for heart disease diagnosis. Phys A: Stat Mech Appl 482:796–807
Subanya B, Rajalaxmi RR (2014) Feature selection using Artificial Bee Colony for cardiovascular disease classification. In: 2014 International Conference on Electronics and Communication Systems (ICECS), pp 1–6
Suresh P, Ananda Raj MD (2018) Study and analysis of prediction model for heart disease: an optimization approach using genetic algorithm. Int J Pure Appl Math 119:5323–5336
Vivekanandan T, Iyengar NCSN (2017) Optimal feature selection using a modified differential evolution algorithm and its effectiveness for prediction of heart disease. Comput Biol Med 90:125–136
Voulodimos A, Doulamis N, Doulamis A, Protopapadakis E (2018) Deep learning for computer vision: a brief review. Comput Intell Neurosci 2018:1–13
Wang Z, Zhu Y, Li D, Yin Y, Zhang J (2020) Feature rearrangement based deep learning system for predicting heart failure mortality. Comput Methods Prog Biomed 191:105383–105383
Yazid MHBA, Talib MS, Satria MH (2019) Flower pollination neural network for heart disease classification. In: IOP Conference Series: Materials Science and Engineering, pp 012072–012072
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
El-Shafiey, M.G., Hagag, A., El-Dahshan, ES.A. et al. A hybrid GA and PSO optimized approach for heart-disease prediction based on random forest. Multimed Tools Appl 81, 18155–18179 (2022). https://doi.org/10.1007/s11042-022-12425-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-022-12425-x