Abstract
In this review article, we present the major insights from and challenges faced in the acquisition, analysis and modeling of astrocyte calcium activity, aiming at bridging the gap between those fields to crack the complex astrocyte “Calcium Code”. We then propose strategies to reinforce interdisciplinary collaborative projects to unravel astrocyte function in health and disease.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
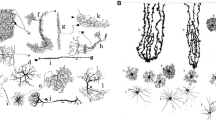
Astrocytes, the most abundant non-neuronal cells of the nervous system, are essential to brain function, from synaptogenesis and neurotransmission to higher brain functions such as memory and learning (Verkhratsky and Nedergaard 2018). Those functions of astrocytes are altered in various brain diseases such as epilepsy, brain tumors, neurodegenerative diseases, Down syndrome, major depressive disorder and schizophrenia (Verkhratsky and Nedergaard 2018). Astrocytes notably respond to stimuli with transient elevations in cytosolic calcium concentration, referred to as calcium signals. Those calcium signals are essential to brain function and are altered in various brain diseases (Verkhratsky and Nedergaard 2018; Semyanov et al. 2020). Importantly, astrocyte calcium signals can trigger the release of molecules referred to as gliotransmitters that modulate neuronal communication at synapses (for recent reviews on gliotransmission and the associated controversies, see (Savtchouk and Volterra 2018; Fiacco and McCarthy 2018)). Better understanding astrocyte physiology and the communication between astrocytes and other cells of the central nervous system thus rely on our ability to make sense of those calcium signals. Astrocyte calcium signals are characterized by diverse spatial (from microdomains to signals spreading within astrocyte networks) and temporal characteristics (from hundreds of milliseconds to tens of seconds) (Semyanov et al. 2020). The majority of those signals occur in fine astrocyte compartments (50-200 nm), referred to as processes, that account for as much as 80 % of the volume of an astrocyte, yet cannot be resolved by diffraction-limited light microscopy (Semyanov et al. 2020; Bindocci et al. 2017) (see Fig. 1). This strongly hinders our ability to characterize astrocyte calcium activity, from the molecular pathways involved to the quantification of the spatio-temporal properties of the signals. Consequently, the functions of the various signals observed, referred to as the astrocyte “Calcium Code”, remain unclear. Better characterizing astrocyte activity concomitantly with the activity of other brain cells will be essential to unravel the roles of astrocyte calcium signals in brain function (Adamsky et al. 2018). Please refer to the review (Semyanov et al. 2020) for more details on the current challenges associated with the study of calcium signals in astrocytes.
In this review article, we highlight the importance of reinforcing interdisciplinary collaborations to crack the astrocyte “Calcium Code”, with a focus on the characterization of the properties of astrocyte calcium signals. We present the major insights from and challenges faced in data acquisition, analysis and modeling of astrocyte calcium activity and propose strategies to facilitate and strengthen collaborations between these fields, which are essential to unravel the functions of astrocyte calcium signals in health and disease.
Acquisition of Astrocyte Calcium Signals
Data acquisition is the first step to characterize astrocyte calcium activity. In this section, we present a brief overview of the tools that are available for imaging astrocyte calcium signals, both in slices and in vivo. We further highlight the insights, challenges and perspectives associated with measuring calcium signals in astrocytes.
Imaging Tools for Astrocyte Calcium Acquisition
The development of calcium indicators, which change their fluorescence properties when binding to calcium ions, allowed neuroscientists to study astrocyte calcium activity. Numerous indicators exist, characterized by diverse kinetics and diffusion properties, so that they should be chosen carefully. In the early days, chemical calcium dyes, such as Fluo-4 or Oregon Green BAPTA, were commonly used (Fellin et al. 2004; Fiacco and McCarthy 2004; Hirase et al. 2004; Parri et al. 2001; Perea and Araque 2005; Serrano et al. 2006). One of the main caveats of these chemical sensors is the low signal-to-background noise ratio of the resulting signals, which only allows visualizing calcium signals in the soma and the main thick branches of astrocytes (see Fig. 1), unless loaded through a patch-clamp recording pipette and visualized with high-resolution microscopy (Di Castro et al. 2011; Panatier et al. 2011; Min and Nevian 2012). More recently, the development of genetically encoded calcium indicators (GECIs) (Berlin et al. 2015) has considerably improved our understanding of astrocyte calcium dynamics. Various GECIs have been developed in the last years that can be imaged by different tools, for precise or wide imaging at cellular or subcellular levels (Agarwal et al. 2017; Durkee and Araque 2019; Shigetomi et al. 2013; Shigetomi et al. 2010; Serrat et al. 2021). GECIs have several advantages compared to classical calcium dyes. First, they can be easily targeted to be expressed specifically in astrocytes. Moreover, they provide a higher signal-to-background noise ratio compared to classical calcium dyes and diffuse better into the fine processes. Additionally, GECIs can be expressed in live organisms, thus allowing in vivo calcium imaging in anesthetized (Lines et al. 2020; Poskanzer and Yuste 2016; Stobart et al. 2018; Serrat et al. 2021), awake head-fixed (Agarwal et al. 2017; Paukert et al. 2014; Srinivasan et al. 2016; Stobart et al. 2018) or freely moving mice during consecutive behavioral sessions (Corkrum et al. 2020; Paukert et al. 2014; Qin et al. 2020). While many GECIs have been designed in the last years for neurons, only a few are available to target astrocytes specifically (reviewed in (Lohr et al. 2021)). These GECIs have different spectral, temporal and spatial properties that make them suitable for specific experimental applications (Tong et al. 2013). Importantly, they yield calcium signals with different spatio-temporal properties that may not be comparable and may be difficult to analyze with certain software (see “Analysis of Astrocyte Calcium Signals”).
Astrocytes display most of their activity in their fine processes. The majority of those signals are spatially restricted, forming so-called microdomains, and display strikingly diverse spatio-temporal properties (Khakh and McCarthy 2015). Understanding the physiological relevance of those calcium signals requires powerful imaging techniques that can be used in combination with complementary methods to manipulate astrocyte and neuronal activity, such as electrophysiology, optogenetics, pharmacology and behavioral tests. Notably, because of the small size of astrocyte processes, high-resolution microscopy is needed to obtain a thorough view of the astrocyte calcium activity. Both confocal and two-photon microscopy are good options for imaging astrocyte calcium activity because these setups are generally compatible with other techniques, allowing for the study of calcium signals at the tripartite synapse level in slices and in vivo in anesthetized (Poskanzer and Yuste 2016; Stobart et al. 2018; Lines et al. 2020; Serrat et al. 2021) or awake head-fixed mice (Paukert et al. 2014; Agarwal et al. 2017; Stobart et al. 2018; Srinivasan et al. 2016). Light sheet fluorescence microscopy (LSFM) and Lattice LSFM are novel imaging techniques that allow fast 3D scanning with low phototoxicity and a resolution comparable to confocal microscopy (Chen et al. 2014; Ducros et al. 2019). Therefore, those techniques are excellent imaging options for experiments in brain slices. Please refer to Table 1 for an overview of optical resolution, phototoxicity/photobleaching, and compatibility of the different imaging techniques. High-resolution microscopy allows recording calcium signals at a high acquisition speed (in the order of ms), but its spatial resolution is limited by diffraction (x-y: 0.2–0.3 \(\mu\)m and z: 0.5 \(\mu\)m at best) and, thus, does not allow visualizing fine processes in detail. Recent studies have used super-resolution microscopy such as stimulated emission depletion (STED) and stochastic optical reconstruction microscopy (STORM) to study astrocyte morphology at the tripartite synapse level in live tissue (Panatier et al. 2014; Heller et al. 2020; Arizono et al. 2020). STED microscopy has revealed that the complex spongiform morphology of astrocyte processes contains functionally isolated nanostructures that are characterized by spatially restricted calcium signals (Arizono et al. 2020). Currently, because of their low acquisition speed and high laser intensity, which induces high photobleaching of calcium sensors, super-resolution microscopy techniques cannot be used to perform calcium imaging. Thus, in the aforementioned study, calcium signals were acquired using high-resolution microscopy and were then mapped onto super-resolution structural images. Super-resolution imaging requires powerful computational tools, both for acquisition and analysis, which are not broadly available in the experimental community (in terms of knowledge, software and hardware) and emphasizes the need to establish collaborations between experimental and computer scientists.
The Need for Interdisciplinary Approaches
It is important to keep in mind that experimental approaches have inherent limitations. First, calcium indicators are calcium buffers. Therefore, calcium indicators compete with calcium binding sites in the cell, altering calcium signals and the normal functioning of the cell. Second, the spatial and temporal characteristics of the measured signals are constrained by the imaging technique as well as the kinetics of the calcium indicator used. It is thus possible that some faster or smaller calcium signals than those currently reported exist in astrocytes that cannot be detected by the calcium imaging tools that are currently available. Importantly, this effect can be amplified during 3D scanning for calcium signals that are faster than the z-scanning time. Lastly, experimental manipulations, such as using a knock-out mouse line or bath applying drugs, can have unexpected off-site effects that can impact the results, making it difficult to extract causal relationships between the experimental manipulation and the obtained results. Collaborative work with computational scientists is essential to build mechanistic models to go beyond those limitations. For example, models have been essential to characterize the effect of the concentration, kinetics and diffusion coefficient of calcium buffers, such as calcium indicators, on calcium dynamics (Matthews and Dietrich 2015; Schwaller 2010). Models can thus be used to predict the free calcium signals that would occur in the absence of indicators. Further, models can measure in silico calcium signals at very high spatial and temporal resolution (depending on the method used, see "Modeling Astrocyte Calcium Signals"), thus predicting the range of calcium signals that could not be resolved experimentally.
Analysis of Astrocyte Calcium Signals
In the quest of characterizing astrocyte calcium signals, the key role of analysis is to provide tools to experimentalists and modelers to process their data, of increasing size and complexity. Statistical as well as advanced computational image analysis tools are thus needed. In this section, we focus on the analysis of calcium images, which is meant to quantify what is observed, i.e., to extract meaningful information or measurements from images. In particular, we emphasize the importance of developing computational image analysis tools dedicated to the quantification of astrocyte calcium signals, and the challenges to get there.
Image Analysis to Characterize Astrocyte Calcium Signals
Decoding the astrocyte “Calcium Code” involves the characterization of the spatio-temporal dynamics of astrocyte calcium signals. Computational image analysis tools aim at accurately detecting all astrocyte calcium signals in a sequence of microscopy images and, for each signal, extracting its dynamical and spatial features, such as its amplitude, its duration, its trajectory, its propagation speed, the location from which it originates and its volume. Various information, such as the number of calcium signals in a specified region or cell, their frequency at a position, and the different types of signals induced by a stimulus, can be deduced from those measurements.
From an image analysis point of view, reaching this ideal of output information requires preprocessing steps (e.g., denoising, deconvolution, motion correction) as well as the detection, the segmentation and the quantification of the calcium signals in a sequence of microscopy images, which is very challenging due to the complex nature of these signals. First, calcium signals are characterized by various durations (from milliseconds to tens of seconds), frequencies and signal-to-noise ratios. Second, their spatial spreads vary from microdomains to signals that propagate within the astrocyte in regions of various sizes and shapes. Third, they can overlap in space and time (Srinivasan et al. 2015). As most signals occur in fine astrocyte processes that cannot be fully resolved by diffraction-limited light microscopy techniques, image analysis methods cannot rely on morphological criteria to detect calcium signals, which also complexifies their quantification. In addition, the developed image analysis methods should ideally operate across data with different spatial scales, taken in vivo or in vitro, and acquired with different imaging techniques.
Lack of Computational Image Analysis Tools Adapted to the Complexity and Diversity of the Data
Recently, several image analysis algorithms have been developed to quantify astrocyte calcium signals in 2D+time microscopy images. Among them, we can cite GECIquant (Venugopal et al. 2019), CaSCaDe (Agarwal et al. 2017), FASP (Wang et al. 2017), AQuA (Wang et al. 2019) and, more recently, Begonia (Bjørnstad et al. 2021) and Astral (Dzyubenko et al. 2021). Most of these methods are ROI-based approaches (ROI: region of interest), meaning that calcium signals are analyzed through fixed spatial boundaries in the image. As the spatial spread of calcium signals can vary over time and become larger than or get out of the ROI, those approaches can lead to inaccurate or partial detection of the signals. To solve this issue, event-based algorithms have been developed, such as AQuA (Wang et al. 2019). For more details about these algorithms (e.g., analysis approach and outputs), please refer to the dedicated section in the review article from (Lia et al. 2021).
The aforementioned analysis tools have considerably improved the detection and characterization of astrocyte calcium signals. Yet, their use can be restricted, either because they are not adapted to the diversity of acquisition modes and calcium indicators (see “Acquisition of Astrocyte Calcium Signals”) or because they are not open-source or not user-friendly (Carpenter et al. 2012). This can constrain some neuroscientists to implement “in-house” analysis pipelines, which is time-consuming and less reproducible, or to use tools that were initially developed for neuronal calcium imaging analysis, such as CaImAn (Giovannucci et al. 2019), Suite2P (Pachitariu et al. 2017) and LC_Pro (Francis et al. 2012). This latter approach is not optimal as astrocytes differ from neurons in many ways. For example, they have a different morphology. Further, notably because astrocytes are not polarized cells, their calcium signals display different spatio-temporal properties than the ones of neurons.
The continuous scientific and technical advances in calcium imaging will always call for new and adapted image analysis algorithms. Until now, most of the quantification of calcium signals has been performed on 2D+time fluorescence microscopy data. The recent emergence of 3D+time imaging techniques gives access to new and major structural and dynamical information, such as the number of calcium signals occurring in an entire astrocyte volume, their synchronization, their trajectory and the location from which they originate (Bindocci et al. 2017). To the best of our knowledge, there is currently no image analysis tool to detect, segment and quantify astrocyte calcium signals in 3D+time microscopy images.
Challenges Hindering the Development of 3D+time Image Analysis Tools
The main reason why the quantification of astrocyte calcium signals has been so far restricted to 2D+time images is because of the trade-off between temporal and spatial resolution in microscopy techniques. The access to a refined 3D imaging of the dynamical behavior of calcium signals in astrocytes is quite recent, owing to the emergence of microscopes enabling a high 3D spatio-temporal resolution with low phototoxicity (e.g., lattice light sheet fluorescence microscopy (LSFM) (Chen et al. 2014; Ducros et al. 2019) and of genetically encoded calcium indicators (GECIs) (Berlin et al. 2015). Despite these scientific and technical advances, the development of 3D+time image analysis tools tailored for the astrocyte calcium activity is not straightforward and calls for new quantitative analysis algorithms with new constraints and challenges. First, a key challenge in the development of 3D+time image analysis tools is the memory and computational costs required to process large 3D+time data. To give the reader an idea, the equivalent of one hour of acquisition of Lattice LSFM data represents about 1 To of data. Importantly, the developed analysis tools should be accessible and thus ideally be able to run on standard desktop computers. To tackle this challenge, ingenious solutions for image processing are needed such as using data-dimensionality reduction techniques. Second, and more critical, reliable and large amounts of labeled data are not available. Such datasets are crucial to evaluate image analysis tools and to train data-driven tools, which are increasingly common with the emergence of deep learning in biological image analysis (Meijering 2020). Manually annotating 3D time-lapse images is a tedious task—mainly because of the complex visualization in 4D space—that cannot be performed reliably. There is a significant intra- and inter-experimenter variability. There is currently a major lack of annotations of astrocyte calcium activity images. Note that this is also true for other datasets of 3D images in live tissue (Yakimovich et al. 2021). For all of those reasons, a common and promising approach is to use realistic synthetic datasets with known ground-truths (i.e., all morphological and dynamical properties are known and controlled) to train and quantitatively assess the performance of analysis software. This highlights the need for developing models and simulators that are able to mimic real image sequences.
Need for Public Realistic Synthetic Datasets: Join the Forces!
To solve the difficulty to reliably label calcium signals in microscopy images, a promising approach consists in generating 3D+time synthetic datasets that realistically depict astrocyte calcium signals observed in real microscopy images. To be as realistic as possible, the simulation should be driven by a biophysical model that describes the calcium signals at the nanoscale, which requires close collaboration between image analysts, modelers (see “Modeling Astrocyte Calcium Signals”) and experimentalists (see “Acquisition of Astrocyte Calcium Signals”). For instance, a recent interdisciplinary collaboration (Badoual et al. 2021) has resulted in the creation of a simulator to generate realistic sequences of 3D lattice LSFM images depicting calcium activity in the sponge-like network of astrocyte processes by integrating a simplified version of the kinetic model developed by (Denizot et al. 2019). In addition to hopefully opening the door to the deployment of 3D+time image analysis tools to quantify astrocyte calcium activity, these simulators could also help modelers tuning their models and the parameters in a faster way than using computational simulations, which are often time and computationally expensive. A major challenge to develop such simulators is the complexity of evaluating the similarity between the generated synthetic images and real images. Implementing rigorous methods to evaluate synthetic astrocyte calcium images will thus be essential to ensure the success of this approach. Note that these synthetic data are essential to guide analysts in the development of their algorithms, but final qualitative validation on real images is still required.
Modeling Astrocyte Calcium Signals
Models correspond to simplifications that describe relevant parameters of a system of interest (its elements, their states and their interactions), allowing for better quantification, visualization, and understanding of the system. The famous quotation from George Box, “All models are wrong but some are useful” (Box 1980), highlights that models are incomplete representations of the system as a whole, yet provide crucial insights into the system’s behavior and dynamics. Such insights would not be grasped by a model as complex as the system of interest itself.
Depending on the question and hypothesis that emerge from experimental data, modelers choose different approaches and toolkits (see Table 2). For example, models studying calcium activity in microdomains will need a higher spatial resolution than models of somatic signals. Further, the modeling approaches that are well-suited to study calcium microdomains, such as particle-based methods (see (Denizot et al. 2020) for a review), are more accurate but extremely demanding in terms of computational power and simulation time. Simulating hundreds of seconds of calcium activity in a fine astrocyte process (e.g., 1 \(\mu\)m long, 200 nm in radius) can take days to compute, so that using those tools to simulate signals in a whole cell or in a network of cells is currently unfeasible. Please note that the computation time to simulate, e.g., 1 millisecond of calcium activity varies not only depending on the modeling technique used, but also on the computational resources available in each laboratory, on the volume and number of reactions modeled as well as the simulation time. To learn more about the different approaches that can be used to model reactions, their insights and limitations, please refer to dedicated reviews (Burrage et al. 2011; Denizot et al. 2020). The goal of this section is not to present an exhaustive list of astrocyte models (see (Denizot et al. 2020; Manninen et al. 2018; Oschmann et al. 2017)), to review existing models of calcium signaling (reviewed in (Dupont et al. 2011; Dupont et al. 2016; Dupont and Croisier 2010; Dupont and Sneyd 2017; Rüdiger 2014; Rüdiger and Shuai 2019)), or to present a detailed list of modeling tools to model calcium signals (Dupont et al. 2016; Blackwell 2013). Rather, we emphasize the key insights that can be gained from models of astrocyte calcium activity as well as the challenges that computational neuroscientists are currently facing.
Insights from Modeling into Biological Processes
Mathematical and computational models are powerful tools that provide new insights in the mechanisms that regulate calcium activity in astrocytes and generate testable predictions. First, models can be used to conduct in silico experiments that are time-consuming or unfeasible experimentally. Models have for example been used to finely tune the spatial distribution of calcium channels (molecules that, when open, result in a calcium influx into the cytosol, forming a calcium signal) within the cell and explore its impact on astrocyte activity (Denizot et al. 2019; Savtchenko et al. 2018). Moreover, models can be used to generate realistic datasets that can be used to train tools developed to characterize the system’s behavior (see “Analysis of Astrocyte Calcium Signals”) (Badoual et al. 2021). Lastly, computational models are useful to go beyond correlational observations and to propose mechanistic principles that explain experimentally observed data. For example, models have shown the effect of cellular morphology on the compartmentalization of calcium signals in dendritic spines (Bell et al. 2019; Biess et al. 2011; Santamaria et al. 2011; Yasuda 2017) as well as in astrocyte processes (Denizot et al. 2022). For a recent review on the insights gained from computational approaches on astrocyte function as well as strategies to start incorporating astrocyte calcium signals in systems neuroscience to better understand how astrocytes contribute to brain computation, see (Kastanenka et al. 2019). Overall, modeling approaches can provide key insights to astrocyte physiology.
Main Challenges Associated With the Development of Models of Astrocyte Calcium Activity
Computational neuroscientists are facing major challenges to build models of astrocyte calcium activity. First, a lot of data are currently missing or not shared publicly, so that most parameter values used in the astrocyte models that are currently available are derived from data obtained in other cell types. Those data include the concentration and sub-cellular distribution of endogenous buffers, the diffusion coefficient of diffusing molecules involved in calcium dynamics in astrocytes, the distribution of the major calcium channels and pumps in the cell, the dynamic remodeling of the morphology of the cell and of its internal calcium stores in live tissue. Second, the computational cost of simulations increases drastically as the accuracy of the model increases, so that a trade-off often has to be made by the modeler, resulting in models with few reactions or low spatial resolution. Further, some of the currently available models suffer from a lack of availability, replicability, and reproducibility (Manninen et al. 2018). Lastly, models often focus on specific spatio-temporal scales of astrocyte activity. Bridging those models together is critical to better understand the involvement of local calcium signals in higher-level brain functions such as cognition and learning. Building such multi-scale models is challenging but should provide unprecedented insights in the involvement of astrocyte calcium signals in the activity of neural circuits and overall in brain (dys-)function.
Is There such a Thing as a Generic Astrocyte Model?
Although astrocytes share common morphological and biochemical characteristics, they are remarkably heterogeneous. The diversity of astrocyte morphology has been described as early as 100 years ago by Cajal and morphology-based classifications of astrocytes have been proposed (Emsley and Macklis 2006). Astrocyte electrophysiological properties (D’Ambrosio et al. 1998; Nimmerjahn et al. 2009; Takata and Hirase 2008), gene (Doyle et al. 2008; Molofsky et al. 2014; Shah et al. 2016) and protein expression levels (Oberheim et al. 2012) also vary drastically depending on the brain region under study. Those observations suggest that astrocytes are a heterogeneous cell population, questioning the specificities and roles of individual sub-types. For more details, see dedicated reviews on astrocyte heterogeneity (Bayraktar et al. 2015; Haim and Rowitch 2017; Verkhratsky and Nedergaard 2018; Zhou et al. 2019). Whether the diverse functions of astrocytes in the brain rely on molecularly and morphologically distinct sub-populations of astrocytes is still poorly understood, yet crucial to uncover the functions of astrocytes in the healthy and diseased brain. A recent study identified sub-populations of astrocytes that selectively contributed to specific functions such as synaptogenesis and tumor invasion of glioma (John Lin et al. 2017). Incorporating this diversity in astrocyte models by building models of specific sub-populations of astrocytes rather than the currently available generic astrocyte models will be essential to provide insights into the functional implications of the molecular and morphological heterogeneity of astrocytes that have been reported recently.
Need for Interdisciplinary Collaborations to Improve Models of Astrocyte Calcium Activity
Several strategies and perspectives could be developed to go beyond the aforementioned challenges to model astrocyte calcium activity. First, computational neuroscientists would highly benefit from the existence of open-source datasets, which could be used to build and test models. Such datasets are crucial for data-driven modeling practices, which rely on strong iterative collaborative work between experimentalists and computational neuroscientists. Moreover, several good practices and step-by-step modeling guides have been published to ensure the reproducibility of models (Blohm et al. 2020; Kording et al. 2018; Novére et al. 2005). Lastly, initiatives such as the Neuromatch Academy courses and conferences (Achakulvisut et al. 2021; van Viegen et al. 2021) provide an unprecedented opportunity to build an accessible, democratic, inclusive, international and interdisciplinary community aiming at using computational approaches to improve our understanding of brain function.
Perspectives
Astrocytes are cells that display a highly complex activity that is essential to brain function. Characterizing the diverse signals displayed by active astrocytes and understanding their physiological roles, the “Calcium Code”, are the biggest challenges of the field and are crucial to understand the involvement of astrocytes in brain function. In this short review article, we highlighted the different insights that can be gained from each field that studies calcium signals in astrocytes and the major challenges that they are facing. Key challenges that prevent us from making sense of astrocyte calcium activity have arisen from our discussions during our interdisciplinary workshop, hosted by the \(1^{st}\) Virtual Conference of the European Society for Neurochemistry “Future perspectives for European neurochemistry—a young scientist’s conference”, in May 2021, entitled “Let’s join forces—Bridging the gap between experimental, computational and data sciences to disentangle astrocyte calcium activity”. Those challenges include:
-
The development of analysis tools allowing accurate detection and characterization of individual calcium signals in astrocytes are lacking, notably in 3D+time.
-
There is no consensus in key definitions and terminology, which further hinders efficient communication across fields (e.g., calcium microdomain/nanodomain, processes/leaflets, Calcium Code, gliapil/spongiform domain).
-
A lot of data are missing to fully grasp the mechanisms regulating astrocyte calcium signals and their physiological roles. For example, local and regional variability of the expression levels of proteins involved in calcium signaling, both in health and disease, remain to be characterized. The morphology of perisynaptic astrocyte processes and their organelles, together with their dynamical remodeling, also remain to be uncovered in live tissue.
-
Raw data are rarely shared in public repositories. Notably, labeled datasets are needed to evaluate image analysis tools and to train data-driven tools. Providing public access to such datasets has contributed to fast improvements in other fields, such as the development of tools detecting the onset of epileptic seizures (Brinkmann et al. 2016).
-
Interdisciplinary events and projects are rare, which constitutes a major barrier to our efforts to unravel the astrocyte “Calcium Code”. Indeed, scientists from different fields lack opportunities to discuss, share ideas and knowledge. The interactions between fields working on astrocyte calcium signals and opportunities for improvements are highlighted in Fig. 2. We believe that such joint efforts are essential to fully grasp the complex properties and functions of astrocytes.
Reinforcing interdisciplinary collaborations to unravel the astrocyte “Calcium Code”. Left: workflow for the characterization of calcium signals involving the fields of acquisition, analysis and modeling. The raw data acquired by experimentalists include e.g., calcium images, structural images or omics data. Raw data processing by analysts results in dynamical (e.g., duration, trajectory, frequency) and structural characterization (e.g., protein localization, cell morphology) of astrocytes as well as the quantification of protein expression levels, for example. Right: schematic representation of the interactions between the fields. Interactions to reinforce are highlighted in dashed blue lines (4, 5, 6)
Reinforcing interdisciplinary projects, bringing together experts from different fields, will be crucial to ensure our success in cracking the astrocyte “Calcium Code”. Such collaborative projects are still rare in the field, which might result from the high fragmentation of research projects and fields working on astrocyte physiology, often presenting their work in different, highly specialized conferences and journals. We propose initiatives that will facilitate the emergence of new interdisciplinary projects:
-
Agreement on shared definitions and terminology across fields.
-
Sharing datasets, together with all the relevant information on the data acquisition, processing and modeling (if relevant) methods used. This might require the creation of an online platform to store and discuss data on astrocytes.
-
Sharing user-friendly data analysis tools, including providing the code in open-access and the dataset(s) used to facilitate their dissemination to the whole community.
-
Organization of recurrent meetings and events that bring together experts from various fields of expertise.
Because of the complexity of astrocyte morphology and signaling, interdisciplinary projects will be essential to not only crack the astrocyte “Calcium Code”, but also to successfully improve our understanding of astrocyte (patho-)physiology and to propose models of astrocyte function.
Data Availability
Data sharing is not applicable to this article as no datasets were generated or analyzed during this study.
References
Achakulvisut T, Ruangrong T, Mineault P, Vogels TP, Peters MAK, Poirazi P, Rozell C, Wyble B, Goodman DFM, Kording KP (2021) Towards democratizing and automating online conferences: lessons from the neuromatch conferences. Trends Cogn Sci 25(4):265–268. https://doi.org/10.1016/j.tics.2021.01.007
Adamsky A, Kol A, Kreisel T, Doron A, Ozeri-Engelhard N, Melcer T, Refaeli R, Horn H, Regev L, Groysman M, London M, Goshen I (2018) Astrocytic activation generates de novo neuronal potentiation and memory enhancement. Cell 174(1):59–7114. https://doi.org/10.1016/j.cell.2018.05.002
Agarwal A, Wu P-H, Hughes EG, Fukaya M, Tischfield MA, Langseth AJ, Wirtz D, Bergles DE (2017) Transient opening of the mitochondrial permeability transition pore induces microdomain calcium transients in astrocyte processes. Neuron 93(3):587–6057. https://doi.org/10.1016/j.neuron.2016.12.034
Arizono M, Inavalli VVGK, Panatier A, Pfeiffer T, Angibaud J, Levet F, Veer MJTT, Stobart J, Bellocchio L, Mikoshiba K, Marsicano G, Weber B, Oliet SHR, Nägerl UV (2020) Structural basis of astrocytic Ca2+ signals at tripartite synapses. Nat Commun 11(1):1–15. https://doi.org/10.1038/s41467-020-15648-4. Number: 1 Publisher: Nature Publishing Group
Badoual A, Arizono M, Denizot A, Ducros M, Berry H, Nägerl UV, Kervrann C (2021) Simulation of astrocytic calcium dynamics in lattice light sheet microscopy images. In: 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI). pp 135–139. https://doi.org/10.1109/ISBI48211.2021.9433984. ISSN: 1945-8452
Bayraktar OA, Fuentealba LC, Alvarez-Buylla A, Rowitch DH (2015) Astrocyte development and heterogeneity Cold Spring Harb Perspect Biol 7(1):020362. https://doi.org/10.1101/cshperspect.a020362
Bell M, Bartol T, Sejnowski T, Rangamani P (2019) Dendritic spine geometry and spine apparatus organization govern the spatiotemporal dynamics of calcium. J Gen Physiol 151(8):1017–1034. https://doi.org/10.1085/jgp.201812261
Benninger RKP, Piston DW (2013) Two-photon excitation microscopy for the study of living cells and tissues. Current protocols in cell biology / editorial board, Juan S. Bonifacino ... [et al.] 0 4:4–1124. https://doi.org/10.1002/0471143030.cb0411s59
Berlin S, Carroll EC, Newman ZL, Okada HO, Quinn CM, Kallman B, Rockwell NC, Martin SS, Lagarias JC, Isacoff EY (2015) Photoactivatable genetically encoded calcium indicators for targeted neuronal imaging. Nat Methods 12(9):852–858. https://doi.org/10.1038/nmeth.3480
Biess A, Korkotian E, Holcman D (2011) Barriers to diffusion in dendrites and estimation of calcium spread following synaptic inputs. PLoS Comput Biol 7(10):1002182. https://doi.org/10.1371/journal.pcbi.1002182
Bindocci E, Savtchouk I, Liaudet N, Becker D, Carriero G, Volterra A (2017) Three-dimensional Ca2+ imaging advances understanding of astrocyte biology. Science 356(6339):8185. https://doi.org/10.1126/science.aai8185
Bjørnstad DM, Åbjørsbråten KS, Hennestad E, Cunen C, Hermansen GH, Bojarskaite L, Pettersen KH, Vervaeke K, Enger R (2021) Begonia-A two-photon imaging analysis pipeline for astrocytic Ca2+ signals. Front Cell Neurosci 15:176. https://doi.org/10.3389/fncel.2021.681066
Blackwell KT (2013) Approaches and tools for modeling signaling pathways and calcium dynamics in neurons. J Neurosci Methods 220(2):131–140. https://doi.org/10.1016/j.jneumeth.2013.05.008
Blohm G, Kording KP, Schrater PR (2020) A how-to-model guide for neuroscience. eNeuro 7(1). https://doi.org/10.1523/ENEURO.0352-19.2019. Publisher: Society for Neuroscience Section: Research Article: Methods/New Tools
Box GEP (1980) Sampling and Bayes’ Inference in Scientific Modelling and Robustness. Journal of the Royal Statistical Society: Series A (General) 143(4):383–404. https://doi.org/10.2307/2982063
Brazhe AR, Postnov DE, Sosnovtseva O (2018) Astrocyte calcium signaling: interplay between structural and dynamical patterns. Chaos: An Interdisciplinary Journal of Nonlinear Science 28(10):106320. https://doi.org/10.1063/1.5037153
Brinkmann BH, Wagenaar J, Abbot D, Adkins P, Bosshard SC, Chen M, Tieng QM, He J, Muñoz-Almaraz FJ, Botella-Rocamora P, Pardo J, Zamora-Martinez F, Hills M, Wu W, Korshunova I, Cukierski W, Vite C, Patterson EE, Litt B, Worrell GA (2016) Crowdsourcing reproducible seizure forecasting in human and canine epilepsy. Brain 139(6):1713–1722. https://doi.org/10.1093/brain/aww045
Burrage K, Burrage PM, Leier A, Marquez-Lago T, Nicolau DV (2011) Stochastic simulation for spatial modelling of dynamic processes in a living cell. In: Koeppl H, Setti G, di Bernardo M, Densmore D (eds) Design and Analysis of Biomolecular Circuits: Engineering Approaches to Systems and Synthetic Biology. Springer, New York, NY, pp. 43–62. https://doi.org/10.1007/978-1-4419-6766-4_2
Carpenter AE, Kamentsky L, Eliceiri KW (2012) A call for bioimaging software usability. Nat Methods 9(7):666–670. https://doi.org/10.1038/nmeth.2073
Chen B-C, Legant WR, Wang K, Shao L, Milkie DE, Davidson MW, Janetopoulos C, Wu XS, Hammer JA, Liu Z, English BP, Mimori-Kiyosue Y, Romero DP, Ritter AT, Lippincott-Schwartz J, Fritz-Laylin L, Mullins RD, Mitchell DM, Bembenek JN, Reymann A-C, Böhme R, Grill SW, Wang JT, Seydoux G, Tulu US, Kiehart DP, Betzig E (2014) Lattice light-sheet microscopy: Imaging molecules to embryos at high spatiotemporal resolution. Science 346(6208):1257998. https://doi.org/10.1126/science.1257998
Corkrum M, Covelo A, Lines J, Bellocchio L, Pisansky M, Loke K, Quintana R, Rothwell PE, Lujan R, Marsicano G, Martin ED, Thomas MJ, Kofuji P, Araque A (2020) Dopamine-evoked synaptic regulation in the nucleus accumbens requires astrocyte activity. Neuron 105(6):1036–10475. https://doi.org/10.1016/j.neuron.2019.12.026
Cresswel-Clay E, Crock N, Tabak J, Erlebacher G (2018) A compartmental model to investigate local and global Ca2+ dynamics in astrocytes. Front Comput Neurosci 12. https://doi.org/10.3389/fncom.2018.00094
D’Ambrosio R, Wenzel J, Schwartzkroin PA, McKhann GM, Janigro D (1998) Functional specialization and topographic segregation of hippocampal astrocytes. J Neurosci Off J Soc Neurosci 18(12):4425–4438
Denizot A, Arizono M, Nägerl UV, Berry H, De Schutter E (2022) Control of Ca2+ signals by astrocyte nanoscale morphology at tripartite synapses. bioRxiv, 2021–0224432635. https://doi.org/10.1101/2021.02.24.432635
Denizot A, Arizono M, Nägerl UV, Soula H, Berry H (2019) Simulation of calcium signaling in fine astrocytic processes: Effect of spatial properties on spontaneous activity. PLOS Comput Biol 15(8):1006795. https://doi.org/10.1371/journal.pcbi.1006795
Denizot A, Berry H, Venugopal S (2020) Intracellular Calcium Signals in Astrocytes, Computational Modeling of. In: Jaeger D, Jung R (eds) Encyclopedia of Computational Neuroscience. Springer, New York, NY, pp 1–12. https://doi.org/10.1007/978-1-4614-7320-6_100693-1
De Pittá M, Brunel N, Volterra A (2016) Astrocytes: orchestrating synaptic plasticity? Neuroscience 323:43–61. https://doi.org/10.1016/j.neuroscience.2015.04.001
Di Castro MA, Chuquet J, Liaudet N, Bhaukaurally K, Santello M, Bouvier D, Tiret P, Volterra A (2011) Local Ca2+ detection and modulation of synaptic release by astrocytes. Nat Neurosci 14(10):1276–1284. https://doi.org/10.1038/nn.2929
Doyle JP, Dougherty JD, Heiman M, Schmidt EF, Stevens TR, Ma G, Bupp S, Shrestha P, Shah RD, Doughty ML, Gong S, Greengard P, Heintz N (2008) Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell 135(4):749–762. https://doi.org/10.1016/j.cell.2008.10.029
Durkee CA, Araque A (2019) Diversity and specificity of astrocyte-neuron communication. Neuroscience 396:73–78. https://doi.org/10.1016/j.neuroscience.2018.11.010
Ducros M, Getz A, Arizono M, Pecoraro V, Fernandez-Monreal M, Letellier M, Nägerl UV, Choquet D (2019) Lattice light sheet microscopy and photo-stimulation in brain slices. In: Neural Imaging and Sensing 10865:1086508. https://doi.org/10.1117/12.2509467
Dupont G, Combettes L, Bird GS, Putney JW (2011) Calcium oscillations. Cold Spring Harb Perspect Biol 3(3). https://doi.org/10.1101/cshperspect.a004226
Dupont G, Croisier H (2010) Spatiotemporal organization of Ca2+ dynamics: A modeling based approach. HFSP Journal 4(2):43–51. https://doi.org/10.2976/1.3385660
Dupont G, Falcke M, Kirk V, Sneyd J (2016) Models of calcium signalling. Interdiscip Appl Math 43. Springer, Cham. https://doi.org/10.1007/978-3-319-29647-0. http://link.springer.com/10.1007/978-3-319-29647-0
Dupont G, Falcke M, Kirk V, Sneyd J (2016) The calcium toolbox. In: Dupont G, Falcke M, Kirk V, Sneyd J (eds) Models of Calcium Signalling. Interdisciplinary Applied Mathematics. Springer, Cham, pp 29–96. https://doi.org/10.1007/978-3-319-29647-0_2
Dupont G, Sneyd J (2017) Recent developments in models of calcium signalling. Curr Opin Syst Biol 3:15–22. https://doi.org/10.1016/j.coisb.2017.03.002
Dzyubenko E, Prazuch W, Pillath-Eilers M, Polanska J, Hermann DM (2021) Analysing intercellular communication in astrocytic networks using “Astral”. Front Cell Neurosci 15. https://doi.org/10.3389/fncel.2021.689268. Publisher: Frontiers
Emsley J, Macklis J (2006) Astroglial heterogeneity closely reflects the neuronal-defined anatomy of the adult murine CNS. Neuron Glia Biol 2(3). https://doi.org/10.1017/S1740925X06000202
Fellin T, Pascual O, Gobbo S, Pozzan T, Haydon PG, Carmignoto G (2004) Neuronal synchrony mediated by astrocytic glutamate through activation of extrasynaptic NMDA receptors. Neuron 43(5):729–743. https://doi.org/10.1016/j.neuron.2004.08.011
Fiacco TA, McCarthy KD (2004) Intracellular astrocyte calcium waves in situ increase the frequency of spontaneous AMPA receptor currents in CA1 pyramidal neurons. J Neurosci 24(3):722–732. https://doi.org/10.1523/JNEUROSCI.2859-03.2004
Fiacco TA, McCarthy KD (2018) Multiple lines of evidence indicate that gliotransmission goes not occur under physiological conditions. J Neurosci 38(1):3–13. https://doi.org/10.1523/JNEUROSCI.0016-17.2017
Francis M, Qian X, Charbel C, Ledoux J, Parker JC, Taylor MS (2012) Automated region of interest analysis of dynamic Ca2+ signals in image sequences. Am J Physiol Cell Physiol 303(3):236–243. https://doi.org/10.1152/ajpcell.00016.2012. Publisher: American Physiological Society
Giovannucci A, Friedrich J, Gunn P, Kalfon J, Brown BL, Koay SA, Taxidis J, Najafi F, Gauthier JL, Zhou P, Khakh BS, Tank DW, Chklovskii DB, Pnevmatikakis EA (2019) CaImAn an open source tool for scalable calcium imaging data analysis. eLife 8:38173. https://doi.org/10.7554/eLife.38173. Publisher: eLife Sciences Publications, Ltd
Gordleeva SY, Lebedev SA, Rumyantseva MA, Kazantsev VB (2018) Astrocyte as a detector of synchronous events of a neural network. JETP Lett 107(7):440–445. https://doi.org/10.1134/S0021364018070032
Haim LB, Rowitch DH (2017) Functional diversity of astrocytes in neural circuit regulation. Nat Rev Neurosci 18(1):31–41. https://doi.org/10.1038/nrn.2016.159
Heller JP, Odii T, Zheng K, Rusakov DA (2020) Imaging tripartite synapses using super-resolution microscopy. Methods 174:81–90. https://doi.org/10.1016/j.ymeth.2019.05.024
Héja L, Kardos J (2020) NCX activity generates spontaneous Ca2+ oscillations in the astrocytic leaflet microdomain. Cell Calcium 86:102137. https://doi.org/10.1016/j.ceca.2019.102137
Hirase H, Qian L, Bartho P, Buzsáki G (2004) Calcium dynamics of cortical astrocytic networks in vivo. PLoS Biol 2:96. https://doi.org/10.1371/journal.pbio.0020096
Höfer T, Venance L, Giaume C (2002) Control and plasticity of intercellular calcium waves in astrocytes: a modeling approach. J Neurosci 22(12):4850–4859
John Lin C-C, Yu K, Hatcher A, Huang T-W, Lee HK, Carlson J, Weston MC, Chen F, Zhang Y, Zhu W, Mohila CA, Ahmed N, Patel AJ, Arenkiel BR, Noebels JL, Creighton CJ, Deneen B (2017) Identification of diverse astrocyte populations and their malignant analogs. Nat Neurosci 20(3):396–405. https://doi.org/10.1038/nn.4493
Kastanenka KV, Moreno-Bote R, Pittá MD, Perea G, Eraso-Pichot A, Masgrau R, Poskanzer KE, Galea E (2019) A roadmap to integrate astrocytes into Systems Neuroscience. Glia 0(0). https://doi.org/10.1002/glia.23632
Khakh BS, McCarthy KD (2015) Astrocyte calcium signaling: from observations to functions and the challenges therein. Cold Spring Harb Perspect Biol 7(4):020404. https://doi.org/10.1101/cshperspect.a020404
Kording K, Blohm G, Schrater P, Kay K (2018) Appreciating diversity of goals in computational neuroscience. Technical report, OSF Preprints. https://doi.org/10.31219/osf.io/3vy69. type: article. https://osf.io/3vy69/
Kuchibhotla KV, Lattarulo CR, Hyman BT, Bacskai BJ (2009) Synchronous hyperactivity and intercellular calcium waves in astrocytes in Alzheimer mice. Science 323(5918):1211–1215. https://doi.org/10.1126/science.1169096
Lallouette J, De Pittá M, Ben-Jacob E, Berry H (2014) Sparse short-distance connections enhance calcium wave propagation in a 3D model of astrocyte networks. Front Comput Neurosci 8:45. https://doi.org/10.3389/fncom.2014.00045
Lavrentovich M, Hemkin S (2008) A mathematical model of spontaneous calcium (II) oscillations in astrocytes. J Theor Biol 251(4):553–560. https://doi.org/10.1016/j.jtbi.2007.12.011
Lia A, Henriques VJ, Zonta M, Chiavegato A, Carmignoto G, Gómez-Gonzalo M, Losi G (2021) Calcium signals in astrocyte microdomains, a decade of great advances. Front Cell Neurosci 15. https://doi.org/10.3389/fncel.2021.673433. Publisher: Frontiers
Lines J, Martin ED, Kofuji P, Aguilar J, Araque A (2020) Astrocytes modulate sensory-evoked neuronal network activity. Nat Commun 11(1):3689. https://doi.org/10.1038/s41467-020-17536-3
Lohr C, Beiersdorfer A, Fischer T, Hirnet D, Rotermund N, Sauer J, Schulz K, Gee CE (2021) Using genetically encoded calcium indicators to study astrocyte physiology: a field guide. Front Cell Neurosci 0. https://doi.org/10.3389/fncel.2021.690147. Publisher: Frontiers
Manninen T, Havela R, Linne M-L (2018) Computational models for calcium-mediated astrocyte functions. Front Comput Neurosci. https://doi.org/10.3389/fncom.2018.00014
Matthews EA, Dietrich D (2015) Buffer mobility and the regulation of neuronal calcium domains. Front Cell Neurosci 9. https://doi.org/10.3389/fncel.2015.00048
Meijering E (2020) A bird’s-eye view of deep learning in bioimage analysis. Comput Struct Biotechnol J 18:2312–2325. https://doi.org/10.1016/j.csbj.2020.08.003
Min R, Nevian T (2012) Astrocyte signaling controls spike timing-dependent depression at neocortical synapses. Nat Neurosci 15(5):746–753. https://doi.org/10.1038/nn.3075
Molofsky AV, Kelley KW, Tsai H-H, Redmond SA, Chang SM, Madireddy L, Chan JR, Baranzini SE, Ullian EM, Rowitch DH (2014) Astrocyte-encoded positional cues maintain sensorimotor circuit integrity. Nature 509(7499):189–194. https://doi.org/10.1038/nature13161
Nimmerjahn A, Mukamel EA, Schnitzer MJ (2009) Motor behavior activates Bergmann Glial networks. Neuron 62(3):400. https://doi.org/10.1016/j.neuron.2009.03.019
Novére NL, Finney A, Hucka M, Bhalla US, Campagne F, Collado-Vides J, Crampin EJ, Halstead M, Klipp E, Mendes P, Nielsen P, Sauro H, Shapiro B, Snoep JL, Spence HD, Wanner BL (2005) Minimum information requested in the annotation of biochemical models (MIRIAM). Nat Biotechnol 23(12):1509–1515. https://doi.org/10.1038/nbt1156
Oberheim NA, Goldman SA, Nedergaard M (2012) Heterogeneity of astrocytic form and function. Methods Mol Biol (Clifton, N.J.) 814:23–45. https://doi.org/10.1007/978-1-61779-452-0_3
Oschmann F, Berry H, Obermayer K, Lenk K (2017) From in silico astrocyte cell models to neuron-astrocyte network models: a review. Brain Res Bull. https://doi.org/10.1016/j.brainresbull.2017.01.027
Oschmann F, Mergenthaler K, Jungnickel E, Obermayer K (2017) Spatial separation of two different pathways accounting for the generation of calcium signals in astrocytes. PLoS Comput Biol 13(2):1005377. https://doi.org/10.1371/journal.pcbi.1005377
Pachitariu M, Stringer C, Dipoppa M, Schröder S, Rossi LF, Dalgleish H, Carandini M, Harris KD (2017) Suite2p: beyond 10,000 neurons with standard two-photon microscopy. p 061507. https://doi.org/10.1101/061507. Company: Cold Spring Harbor Laboratory Distributor: Cold Spring Harbor Laboratory Label: Cold Spring Harbor Laboratory Section: New Results Type: article. https://www.biorxiv.org/content/10.1101/061507v2
Panatier A, Arizono M, Nägerl UV (2014) Dissecting tripartite synapses with STED microscopy. Phil Trans R Soc B 369(1654):20130597. https://doi.org/10.1098/rstb.2013.0597
Panatier A, Vallée J, Haber M, Murai KK, Lacaille J-C, Robitaille R (2011) Astrocytes are endogenous regulators of basal transmission at central synapses. Cell 146(5):785–798. https://doi.org/10.1016/j.cell.2011.07.022
Parri HR, Gould TM, Crunelli V (2001) Spontaneous astrocytic Ca2+ oscillations in situ drive NMDAR-mediated neuronal excitation. Nat Neurosci 4(8):0801–803. https://doi.org/10.1038/90507
Paukert M, Agarwal A, Cha J, Doze VA, Kang JU, Bergles DE (2014) Norepinephrine controls astroglial responsiveness to local circuit activity. Neuron 82(6):1263–1270. https://doi.org/10.1016/j.neuron.2014.04.038
Perea G, Araque A (2005) Properties of synaptically evoked astrocyte calcium signal reveal synaptic information processing by astrocytes. J Neurosci Off J Soc Neurosci 25(9):2192–2203. https://doi.org/10.1523/JNEUROSCI.3965-04.2005
De Pittá MD, Goldberg M, Volman V, Berry H, Ben-Jacob E (2009) Glutamate regulation of calcium and IP3 oscillating and pulsating dynamics in astrocytes. J Biol Phys 35(4):383–411. https://doi.org/10.1007/s10867-009-9155-y
Poskanzer KE, Yuste R (2016) Astrocytes regulate cortical state switching in vivo. Proc Natl Acad Sci USA 113(19):2675–2684. https://doi.org/10.1073/pnas.1520759113
Postnov DE, Koreshkov RN, Brazhe NA, Brazhe AR, Sosnovtseva OV (2009) Dynamical patterns of calcium signaling in a functional model of neuron-astrocyte networks. J Biol Phys 35(4):425–445. https://doi.org/10.1007/s10867-009-9156-x
Qin H, He W, Yang C, Li J, Jian T, Liang S, Chen T, Feng H, Chen X, Liao X, Zhang K (2020) Monitoring astrocytic Ca2+ activity in freely behaving mice. Front Cell Neurosci 0. https://doi.org/10.3389/fncel.2020.603095. Publisher: Frontiers
Riera J, Hatanaka R, Ozaki T, Kawashima R (2011) Modeling the spontaneous Ca2+ oscillations in astrocytes: Inconsistencies and usefulness. J Integr Neurosci 10(4):439–473. https://doi.org/10.1142/S0219635211002877
Rüdiger S (2014) Stochastic models of intracellular calcium signals. Phys Rep 534(2):39–87. https://doi.org/10.1016/j.physrep.2013.09.002
Rüdiger S, Shuai J (2019) Modeling of stochastic Ca2+ signals. In: De Pittá M, Berry H (eds) Computational Glioscience. Springer Series in Computational Neuroscience. Springer, Cham, pp 91–114. https://doi.org/10.1007/978-3-030-00817-8_4
Santamaria F, Wils S, De Schutter E, Augustine GJ (2011) The diffusional properties of dendrites depend on the density of dendritic spines. Eur J Neurosci 34(4):561–568. https://doi.org/10.1111/j.1460-9568.2011.07785.x
Savtchenko LP, Bard L, Jensen TP, Reynolds JP, Kraev I, Medvedev N, Stewart MG, Henneberger C, Rusakov DA (2018) Disentangling astroglial physiology with a realistic cell model in silico. Nat Commun 9(1):3554. https://doi.org/10.1038/s41467-018-05896-w
Savtchenko LP, Bard L, Jansen TP, Reynolds JP, Kraev I, Medvidov M, Stewart MG, Henneberger C, Rusakov D (2018) Biophysical underpinning of astroglial physiology probed with realistic cell models. bioRxiv, 336974. https://doi.org/10.1101/336974
Savtchouk I, Volterra A (2018) Gliotransmission: beyond black-and-white. J Neurosci 38(1):14–25. https://doi.org/10.1523/JNEUROSCI.0017-17.2017
Schwaller B (2010) Cytosolic Ca2+ buffers. Cold Spring Harb Perspect Biol 2(11):004051. https://doi.org/10.1101/cshperspect.a004051
Semyanov A, Henneberger C, Agarwal A (2020) Making sense of astrocytic calcium signals—from acquisition to interpretation. Nat Rev Neurosci 1–14. https://doi.org/10.1038/s41583-020-0361-8. Publisher: Nature Publishing Group
Serrano A, Haddjeri N, Lacaille J-C, Robitaille R (2006) GABAergic network activation of glial cells underlies hippocampal heterosynaptic depression. J Neurosci 26(20):5370–5382. https://doi.org/10.1523/JNEUROSCI.5255-05.2006
Serrat R, Covelo A, Kouskoff V, Delcasso S, Ruiz-Calvo A, Chenouard N, Stella C, Blancard C, Salin B, Julio-Kalajzic F, Cannich A, Massa F, Varilh M, Deforges S, Robin LM, Stefani DD, Busquets-Garcia A, Gambino F, Beyeler A, Pouvreau S, Marsicano G (2021) Astroglial ER-mitochondria calcium transfer mediates endocannabinoid-dependent synaptic integration. Cell Rep 37(12). https://doi.org/10.1016/j.celrep.2021.110133
Shah S, Lubeck E, Zhou W, Cai L (2016) In situ transcription profiling of single cells reveals spatial organization of cells in the mouse hippocampus. Neuron 92(2):342–357. https://doi.org/10.1016/j.neuron.2016.10.001
Shigetomi E, Bushong EA, Haustein MD, Tong X, Jackson-Weaver O, Kracun S, Xu J, Sofroniew MV, Ellisman MH, Khakh BS (2013) Imaging calcium microdomains within entire astrocyte territories and endfeet with GCaMPs expressed using adeno-associated viruses. J Gen Physiol 141(5):633–647. https://doi.org/10.1085/jgp.201210949
Shigetomi E, Kracun S, Sofroniew MV, Khakh BS (2010) A genetically targeted optical sensor to monitor calcium signals in astrocyte processes. Nat Neurosci 13(6):759–766. https://doi.org/10.1038/nn.2557
Srinivasan R, Huang BS, Venugopal S, Johnston AD, Chai H, Zeng H, Golshani P, Khakh BS (2015) Ca(2+) signaling in astrocytes from Ip3r2(-/-) mice in brain slices and during startle responses in vivo. Nat Neurosci 18(5):708–717. https://doi.org/10.1038/nn.4001
Smith S, Grima R (2018) Spatial stochastic intracellular kinetics: a review of modelling approaches. Bull Math Biol. https://doi.org/10.1007/s11538-018-0443-1
Srinivasan R, Lu TY, Chai H, Xu J, Huang BS, Golshani P, Coppola G, Khakh BS (2016) New transgenic mouse lines for selectively targeting astrocytes and studying calcium signals in astrocyte processes in situ and in vivo. Neuron 92(6):1181–1195. https://doi.org/10.1016/j.neuron.2016.11.030
Stobart JL, Ferrari KD, Barrett MJP, Glück C, Stobart MJ, Zuend M, Weber B (2018) Cortical circuit activity evokes rapid astrocyte calcium signals on a similar timescale to neurons. Neuron 98(4):726–7354. https://doi.org/10.1016/j.neuron.2018.03.050
Stobart JL, Ferrari KD, Barrett MJP, Stobart MJ, Looser ZJ, Saab AS, Weber B (2018) Long-term in vivo calcium imaging of astrocytes reveals distinct cellular compartment responses to sensory stimulation. Cerebral Cortex (New York, N.Y.: 1991) 28(1):184–198. https://doi.org/10.1093/cercor/bhw366
Takata N, Hirase H (2008) Cortical layer 1 and layer 2/3 astrocytes exhibit distinct calcium dynamics in vivo. PLoS One 3(6):2525. https://doi.org/10.1371/journal.pone.0002525
Tong X, Shigetomi E, Looger LL, Khakh BS (2013) Genetically encoded calcium indicators and astrocyte calcium microdomains. The Neuroscientist: A Review Journal Bringing Neurobiology, Neurology and Psychiatry 19(3):274–291. https://doi.org/10.1177/1073858412468794
van Viegen T, Akrami A, Bonnen K, DeWitt E, Hyafil A, Ledmyr H, Lindsay GW, Mineault P, Murray JD, Pitkow X, Puce A, Sedigh-Sarvestani M, Stringer C, Achakulvisut T, Alikarami E, Atay MS, Batty E, Erlich JC, Galbraith BV, Guo Y, Juavinett AL, Krause MR, Li S, Pachitariu M, Straley E, Valeriani D, Vaughan E, Vaziri-Pashkam M, Waskom ML, Blohm G, Kording K, Schrater P, Wyble B, Escola S, Peters MAK (2021) Neuromatch academy: teaching computational neuroscience with global accessibility. Trends Cogn. Sci 25(7):535–538. https://doi.org/10.1016/j.tics.2021.03.018
Verkhratsky A, Nedergaard M (2018) Physiology of astroglia. Physiol Rev 98(1):239–389. https://doi.org/10.1152/physrev.00042.2016
Venugopal S, Srinivasan R, Khakh BS (2019) GECIquant: semi-automated detection and quantification of astrocyte intracellular Ca2+ signals monitored with GCaMP6f. In: De Pittá M, Berry H (eds) Computational Glioscience. Springer Series in Computational Neuroscience. Springer, Cham, pp 455–470. https://doi.org/10.1007/978-3-030-00817-8_17
Wang Y, DelRosso NV, Vaidyanathan TV, Cahill MK, Reitman ME, Pittolo S, Mi X, Yu G, Poskanzer KE (2019) Accurate quantification of astrocyte and neurotransmitter fluorescence dynamics for single-cell and population-level physiology. Nat Neurosci 22(11):1936–1944. https://doi.org/10.1038/s41593-019-0492-2
Wang Y, Shi G, Miller DJ, Wang Y, Wang C, Broussard G, Wang Y, Tian L, Yu G (2017) Automated functional analysis of astrocytes from chronic time-lapse calcium imaging data. Front Neuroinform 11:48. https://doi.org/10.3389/fninf.2017.00048
Yakimovich A, Beaugnon A, Huang Y, Ozkirimli E (2021) Labels in a haystack: approaches beyond supervised learning in biomedical applications. Patterns 2(12). https://doi.org/10.1016/j.patter.2021.100383. Publisher: Elsevier
Yasuda R (2017) Biophysics of biochemical signaling in dendritic spines: implications in synaptic plasticity. Biophys J 113(10):2152–2159. https://doi.org/10.1016/j.bpj.2017.07.029
Zhou B, Zuo Y-X, Jiang R-T (2019) Astrocyte morphology: diversity, plasticity, and role in neurological diseases. CNS Neurosci Ther. https://doi.org/10.1111/cns.13123
Acknowledgements
We thank M. Arizono and C. Vivar Rios for their comments on the manuscript.
Funding
The work from A. Covelo was supported by the Human Frontier Science Program (LT 000827/2019-L3) and the Brain and Behavior Research Foundation. The work from A. Denizot was supported by a JSPS (Japan Society for the Promotion of Science) Standard Postdoctoral Fellowship for Research in Japan (21F21733).
Author information
Authors and Affiliations
Contributions
Ana Covelo wrote the first draft of the data acquisition section, Anaïs Badoual wrote the first draft of the data analysis section, and Audrey Denizot wrote the first draft of the introduction, modeling and perspectives sections. All authors commented on previous versions of the manuscript, read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics Approval and Consent to Participate
Not Applicable.
Consent for Publication
Not Applicable.
Competing Interests
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Covelo, A., Badoual, A. & Denizot, A. Reinforcing Interdisciplinary Collaborations to Unravel the Astrocyte “Calcium Code”. J Mol Neurosci 72, 1443–1455 (2022). https://doi.org/10.1007/s12031-022-02006-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12031-022-02006-w