Abstract
Background
Snap bean, Phaseolus vulgaris L., as a warm-season vegetable, low temperature stress seriously affect the yield and quality. At present, little is known about the genes and molecular regulation mechanism in cold response in snap bean exposed to low temperature.
Objectives
Our objectives were to identify the low temperature response genes in snap bean and to examine differences in the gene response between cold-tolerant and cold-sensitive genotypes.
Methods
We used two highly inbred snap bean lines in this study, the cold-tolerant line ‘120’, and the cold-sensitive line ‘093’. The plants were grown to the three leaf and one heart stage and exposed to 4 °C low temperature. We used RNA sequencing (RNA-seq) to analyze the differences of gene expression.
Results
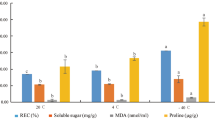
988 and 874 cold-responsive genes were identified in ‘T120 vs CK120’ and ‘T093 vs CK093’ (‘T’ stands for low temperature treatment, and ‘CK’ stands for control at room temperature), respectively. Of these, 555 and 442 genes were unique to cold-stressed lines ‘120’ and ‘093’, respectively compared to the control. Our analysis of these differentially expressed genes indicates that Ca2+, ROS, and hormones act as signaling molecules that play important roles in low temperature response in P. vulgaris. Altering the expression of genes in these signaling pathways activates expression of downstream response genes which can interact with other signaling regulatory networks. This may maintained the balance of ROS and hormones, making line ‘120’ more cold-tolerant than line ‘093’.
Conclusion
Our results provide a preliminarily understanding of the molecular basis of low temperature response in snap bean, and also establish a foundation for the future genetic improvement of cold sensitivity in snap bean by incorporating genes for cold tolerance.







Similar content being viewed by others
References
Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, Van Der Straeten D, Peng J, Harberd NP (2006) Integration of plant responses to environmentally activated phytohormonal signals. Science 311(5757):91–94
Agarwal M, Hao Y, Kapoor A, Dong C, Fujii H, Zheng X, Zhu J (2006) A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J Biol Chem 281:37636–37645
Buskirk HAV, Thomashow MF (2006) Arabidopsis transcription factors regulating cold acclimation. Physiol Plant 126(1):72–80
Chen L, Song Y, Li S, Zhang L, Zou C, Yu D (2012) The role of WRKY transcription factors in plant abiotic stresses. Biochim Biophys Acta 1819(2):128
Cheng C, Yun KY, Ressom HW, Mohanty B, Bajic VB, Jia Y, Yun SJ, Reyes BG (2007) An early response regulatory cluster induced by low temperature and hydrogen peroxide in seedlings of chilling-tolerant japonica rice. BMC Genom 8(1):175
Chinnusamy V, Zhu J, Zhu JK (2007) Cold stress regulation of gene expression in plants. Trends Plant Sci 12(10):451
Craig W, Lenzi P, Scotti N, Craig W, Lenzi P, Scotti N, Palma MD, Saggese P, Carbone V, Curran NM, Magee AM, Medgyesy P, Kavanagh TA, Dix PJ, Grillo S, Cardi T (2008) Transplastomic tobacco plants expressing a fatty acid desaturase gene exhibit altered fatty acid profiles and improved cold tolerance. Transgenic Res 17(5):769–782
Dong MA, Farre EM, Thomashow MF (2011) Circadian clock-associated 1 and late elongated hypocotyl regulate expression of the C-repeat binding factor (CBF) pathway in Arabidopsis. Proc Natl Acad Sci 108(17):7241–7246
Elssa P, Swaleha T, Barik SR, Durga PM, Deepak KN, Shakti PM, Sujata D, Sharat KP (2017) Genome-wide association mapping reveals multiple QTLs governing tolerance response for seedling stage chilling stress in indica rice. Front Plant Sci 8:552
Fowler S, Thomashow MF (2002) Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14:1675–1690
Gilmour SJ, Zarka DG, Stockinger EJ, Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF (1998) Low temperature regulation of the, Arabidopsis, CBF family of AP2 transcriptional activators as an early step in cold-induced, COR gene expression. Plant J 16(4):10
Guo W, Jin L, Miao Y, He X, Hu Q, Guo K, Zhu L, Zhang X (2016) An ethylene response-related factor, GbERF1-like, from Gossypium barbadense improves resistance to Verticillium dahlia via activating lignin synthesis. Plant Mol Biol 91(3):305–318
Habben JE, Bao X, Bate NJ, DeBruin JL, Dolan D, Hasegawa D, Helentjaris TG, Lafitte RH, Lovan N, Mo H, Reimann K, Schussler JR (2014) Transgenic alteration of ethylene biosynthesis increases grain yield in maize under field drought-stress conditions. Plant Biotechnol J 12(6):685–693
Hannah MA, Heyer AG, Hincha DK (2005) A global survey of gene regulation during cold acclimation in Arabidopsis thaliana. PLoS Genet 1(2):e26
Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF (1998) Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280:104–106
Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y (2007) KEGG for linking genomes to life and the environment. Nucleic Acids Res 36:D480–D484
Laudencia-Chingcuanco D, Ganeshan S, You F, Fowler B, Chibbar R, Anderson O (2011) Genome-wide gene expression analysis supports a developmental model of low temperature tolerance gene regulation in wheat (Triticum aestivum L.). BMC Genom 12(1):299
Lee CM, Thomashow MF (2012) Photoperiodic regulation of the C-repeat binding factor (CBF) cold acclimation pathway and freezing tolerance in Arabidopsis thaliana. Proc Natl Acad Sci USA 109:15054–15059
Lee BH, Henderson DA, Zhu JK (2005) The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell 17(11):3155–3175
Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozakib K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-emperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10(8):1391–1406
Liu H, Ouyang B, Zhang J, Wang T, Li H, Zhang Y, Yu C, Ye Z (2012) Differential modulation of photosynthesis, signaling, and transcriptional regulation between tolerant and sensitive tomato genotypes under cold stress. PLOS One. https://doi.org/10.1371/journal.pone.0050785
Livak (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2 (−Delta Delta C (T)) method. Methods 25(4):402–408
Mittler R, Vanderauwera S, Suzuki N, Miller G, Tognetti VB, Vandepoele K, Gollery M, Shulaev V, Breusegem FV (2011) ROS signaling: the new wave? Trends Plant Sci 16(6):309
Mulugeta A, Beneberu S, Somashekhar P, Raghuveer S, Wayne W, Bharat S (2014) Common bean germplasm diversity study for cold tolerance in ethiopia. Am J Plant Sci 5:1842–1850
Nakamichi N, Kusano M, Fukushima A, Nakamichi N, Kusano M, Fukushima A, Kita M, Ito S, Yamashino T, Saito K, Sakakibara H, Mizuno T (2009) Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol 50(3):447–462
Oono Y, Seki M, Satou M, Kei I, Kenji A, Tetsuya S, Miki F, Kazuko Y, Kazuo S (2006) Monitoring expression profiles of Arabidopsis genes during cold acclimation and deacclimation using DNA microarrays. Funct Integr Genom 6(3):212–234
Park MR, Yun KY, Mohanty B, Herath V, Xu F, Wijaya E, Bajic V, Yun S, Reyes B (2010) Supra-optimal expression of the cold-regulated OsMyb4 transcription factor in transgenic rice changes the complexity of transcriptional network with major effects on stress tolerance and panicle development. Plant Cell Environ 33(12):2209–2230
Peleg Z, Blumwald E (2011) Hormone balance and abiotic stress tolerance in crop plants. Curr Opin Plant Biol 14(3):290–295
Puranik S, Sahu PP, Srivastava PS, Prem SS, Manoj P (2012) NAC proteins: regulation and role in stress tolerance. Trends Plant Sci 17:369–381
Qin F, Shinozaki K, Yamaguchi-Shinozaki K (2011) Achievements and challenges in understanding plant abiotic stress responses and tolerance. Plant Cell Physiol 52(9):1569–1582
Reddy AS, Ali GS, Celesnik H, Day IS (2011) Coping with stresses: roles of calcium- and calcium/calmodulin-regulated gene expression. Plant Cell 23:2010–2032
Saavedra X, Modrego A, Rodríguez D, Mary P, Luis S, Gregorio N, Oscar L (2010) The nuclear interactor PYL8/RCAR3 of Fagus sylvatica FsPP2C1 is a positive regulator of abscisic acid signaling in seeds and stress. Plant Physiol 152(1):133–150
Seo PJ, Park MJ, Lim MH, Kim SG, Lee M, Baldwin I (2012) A self-regulatory circuit of CIRCADIAN CLOCK-ASSOCIATED1 underlies the circadian clock regulation of temperature responses in Arabidopsis. Plant Cell 24(9):2427–2442
Shi Y, Tian S, Hou L, Huang X, Zhang X, Guo H (2012) Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis. Plant Cell 24(6):2578–2595
Stockinger EJ, Gilmour SJ, Thomashow MF (1997) Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci 94(3):1035–1040
Sun JQ, Jiang HL, Li CY (2011) Systemin/Jasmonate-mediated systemic defense signaling in tomato. Mol Plant 4:607–615
Sunkar R, Li YF, Jagadeeswaran G (2012) Functions of MicroRNAs in plant stress responses. Trends Plant Sci 17:196–203
Suzuki N, Koussevitzky S, Mittler R, Miller G (2012) ROS and redox signaling in the response of plants to abiotic stress. Plant Cell Environ 35:259–270
Theocharis A, Clément Christophe, Barka EA (2012) Physiological and molecular changes in plants grown at low temperatures. Planta 235(6):1091–1105
Wang X, Shan X, Wu Y, Su S, Li S, Liu H, Han J, Xue C, Yuan Y (2016) iTRAQ-based quantitative proteomic analysis reveals new metabolic pathways responding to chilling stress in maize seedlings. J Proteom 146:14–24
Winfield MO, Lu C, Wilson ID, Coghill JA, Edwards KJ (2010) Plant responses to cold: transcriptome analysis of wheat. Plant Biotechnol J 8:749–771
Zhai H, Bai X, Zhu Y, Li Y, Cai H, Ji W, Ji Z, Liu X, Liu X, Li J (2010) A single-repeat R3-MYB transcription factor MYBC1 negatively regulates freezing tolerance in Arabidopsis. Biochem Biophys Res Commun 394(4):1023
Zhang Z, Huang R (2010) Enhanced tolerance to freezing in tobacco and tomato overexpressing transcription factor TERF2/LeERF2 is modulated by ethylene biosynthesis. Plant Mol Biol 73:241–249
Zhang T, Zhao X, Wang W, Yajiao P, Liyu H, Xiaoyue L, Ying Z, Linghua Z, Daichang Y, Binying F (2012) Comparative transcriptome profiling of chilling stress responsiveness in two contrasting rice genotypes. PLoS One 7:e43274
Zhu J, Shi H, Lee BH, Damsz B, Cheng S, Stirm V, Zhu JK, Paul MH, Ray AB (2004) An Arabidopsis homeodomain transcription factor gene, HOS9, mediates cold tolerance through a CBF-independent pathway. Proc Natl Acad Sci 101(26):9873–9878
Funding
Financial support was provided by the China Postdoctoral Science Foundation (Grant number 2018M631973); the Basic scientific research operating expenses of provincial College in Heilongjiang province (Grant numbers KJCXYB201707 and KJCXYB201705); the National Natural Science Funds of China (Grant number 31771869); the Applied technology research and development plan of Heilongjiang province (Grant number GY2019YF0059 ).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liu, C., Yang, X., Yan, Z. et al. Analysis of differential gene expression in cold-tolerant vs. cold-sensitive varieties of snap bean (Phaseolus vulgaris L.) in response to low temperature stress. Genes Genom 41, 1445–1455 (2019). https://doi.org/10.1007/s13258-019-00870-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-019-00870-2