Abstract
We describe a method to automatically predict radiological scores in spinal Magnetic Resonance Images (MRIs). Furthermore, we also identify and localize the pathologies that are the reasons for these scores. We term these pathological regions the “evidence hotspots”. Our contributions are two-fold: (i) a Convolutional Neural Network (CNN) architecture and training scheme to predict multiple radiological scores on multiple slice sagittal MRIs. The scheme uses multi-task CNN training with augmentation, and handles the class imbalance common in medical classification tasks. (ii) the prediction of a heat-map of evidence hotspots for each score. For both of these, all that is required for training is the class label of the disc or vertebrae, no stronger supervision (such as slice labels) is needed. We report state-of-the-art and near-human performances across multiple radiological scorings including: Pfirrmann grading, disc narrowing, endplate defects, and marrow changes.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Automated detection and localization of abnormalities in medical images is an important task to support clinical decision making across many medical specialties. However, to date, many such techniques have relied on strong supervision for their development; yet obtaining sufficiently large, accurately delineated datasets in medical imaging can be prohibitively expensive due to limits in clinical time. In contrast, scores or even indicators relating to the presence or absence of pathology are more readily available. It is therefore desirable to develop techniques that can learn to predict radiological scores and localise them from weakly labelled training data i.e. (disc-volume) supervision.
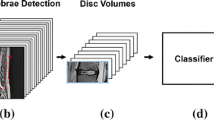
In this paper, we propose a CNN-based framework to classify and qualitatively localize multiple abnormalities in T2 weighted sagittal lumbar MRIs. The method is trained, validated and tested on a dataset of 2009 patients (12018 individual spine disc volumes). The localizations are achieved implicitly through training the network for the classification tasks, and no other labels are needed apart from the classification labels. Given an input scan, the trained model: (i) predicts six separate labels corresponding to six different radiological scores, and (ii) produces six heat-maps, that localizes possible pathological evidence unique to each task. A simplified view of the overall pipeline is given in Fig. 1.
Pipeline: For each of the six intervertebral discs assessed as inputs, there are six corresponding pairs of outputs in the form of evidence hotspot mappings, and predictions of labels specific to the radiological score. Refer to Table 1 for the distribution. The dimension of each input disc volume is \(112 \times 224 \times 9\), essentially a stack of 9 image slices of the disc, which is also the dimension of the evidence hotspot heat-map.
Why multi-task? Despite their advantages and leading performance, one of the challenges of utilizing CNNs in medical imaging remains their need for large training datasets. We address this in our work through the use of multi-tasking where one architecture serves to address multiple problems and hence reduce the number of free parameters. Since each classification problem is its own unique task, solving them at the same time is akin to multi-tasking. Moreover, clinicians are typically required to assess the state of different anatomical regions within the medical image either because they are relevant to the clinical question or because they are required to assess visible anatomy for so-called incidental findings. Hence, the development of techniques that can predict multiple scores simultaneously is desirable; in our application of interest, each disc and vertebrae can have grades to describe their state of normality or degradation.
Why qualitative localization? We believe that the integration of automated quantitative scores into clinical practice can be aided if the system can highlight the regions within the image that lead to the prediction. However, as mentioned above, the cost of obtaining such image mark-up can be prohibitive. Moreover, it is often difficult for a clinician to identify precisely the voxels that resulted in their opinion. Often it is the overall appearance of an area that leads to the conclusion. To this end, we leverage the ability of CNNs to learn to localize important features of objects when trained for object classification tasks, e.g. [8].
Related work: CNNs have been utilized in medical vision problems in two main ways: (1) using networks pre-trained on a larger normally non-medical dataset of natural images, as features, e.g. to detect pathology in chest X-rays [1], and (2) training a network from scratch, e.g. to detect sclerotic metastases in spinal Computed Tomography (CT) images [11]. In comparison, most research on spinal diagnosis classification has been primarily conducted with handcrafted features or what is now referred to as “shallow” learning e.g. in works concerning radiological scoring of the intervertebral discs [3, 5, 7, 9]. One recent successful example of using CNNs on medical images is a segmentation framework proposed by Ronneberger et al. [10] which overcame the problem of small amount of data through the use of elastic augmentation though requiring strong (pixel-level) supervision. Other methods of visualizing disease regions have been investigated in [12].
2 Classification Framework
The goal is to automatically label each disc and the surrounding vertebrae with a number of radiological scores. Two scores are predicted for each disc, Pfirrmann grading and disc narrowing, and four for each vertebrae: lower and upper endplate defects, and lower and upper marrow changes. The six lumbar discs and vertebrae considered are from T12-L1 to L5-S1.
The method starts by first detecting each disc, and then extracting and normalizing a multi-slice volume around the disc based on the vertebral bodies detection framework by [5, 7]. This volume, which includes the disc and part of the vertebral bodies above and below the disc, is the input to the CNN classifier, which predicts all of the scores simultaneously. As shown in Fig. 2 the networks for the various scores share the first five convolutional layers, and then branch out for each individual score. The architecture is described in Sect. 4 and the network is trained simultaneously to predict all the scores using a multi-task loss function. The reason for this choice, rather than say training an individual CNN tower to predict each score, is that there is more supervision on the common part of the architecture (the shared early convolutional layers). We explore the optimum branch point in the experiments. Note, the CNN does not require any localization or segmentation information for training or inference. As will be seen in the results, we achieve state-of-the-art and near human performance.
Multi-task version of the VGG-M architecture [2] with a branch point after the Conv5 layer resulting in a multi-way classification tasks. The numbers in each layer (for example 96 and \(7 \times 7\) in Conv1) refer to the number of filters and their size, /2 denotes a stride of 2, and the max-pooling window is set to be \(2 \times 2\).
Multi-task loss: Since all of the six tasks are classification problems, we follow the standard practice of training the network by minimising the softmax log-losses of all the tasks. For each task, t, where \(t \in \{1 \dots T\}\), and input image, x, the network outputs a vector y of size \(C_t\) which corresponds to the number of classes in task t. The loss, \(\mathcal {L}_{t}\), of each task over N training images can be defined as \(\mathcal {L}_t = -\sum _{n=1}^{N}\left( y_c(x_n) - \log \sum _{j=1}^{C_t} e^{y_j(x_n)}\right) \) where \(y_j\) is the jth component of the FC8 output, and c is the true class of \(x_n\). Solving the multi-task problem in an end-to-end fashion translates to minimizing the combination of all the losses, \(\mathcal {L} = \sum _{t} \omega _t\mathcal {L}_t \) where \(\omega _t\) is the weight of task t. We find setting \(\omega _t = 1\) for every t works well on our tasks, but it might be beneficial to fine tune \(\omega _t\) for different problems. Setting one weight to 1 and the rest to 0 results in a standard training of a single task. At training time, the loss of task t is only calculated for valid labels i.e. missing labels of task t are ignored. This is extremely beneficial as inputs can possess missing labels in one task but not others.
Class-balanced loss per task: Class imbalance refers to the different number of training examples for each possible outcome of a multi-way classification. Since most of the classification tasks we deal with are imbalanced, we use a class-balanced loss during training. For each task, we reweight the loss such that the combined losses are balanced. To achieve balance in training, class-specific weights are introduced. These weights are determined to be \(\alpha _c = freq(c)^{-1}\), where freq(c) are the class frequency in the training sets for each task. The loss for each task can then be expressed as \(\mathcal {L}_t = \sum _c \alpha _c\ell _t(c)\), where \(\ell _t(c)\) is the component of the loss for class c. This is equivalent to oversampling the minority class but, since our data is multi-labelled, this balance can not be achieved by a trivial solution such as simply oversampling or undersampling each disc.
3 Evidence Hotspots
Here we show that a network trained for a fine-grained classification task can produce a heat-map which pinpoints the region in the image space responsible for the prediction. This map lights up pathological areas, ‘hotspots’ of the prediction, specific to the trained task in the image; the brighter the hotspot, the stronger the evidence for that region to influence the classification. Only the class label is used in training, and no other supervisory information is required.
To achieve this, we modify and extend the saliency method proposed by Simonyan et al. [13], that ranks the influence of each pixel in an image, x, according to its influence on the (unnormalized) class score. The method proceeds by linearizing the relation between a specific output class score and the input pixel \(x_p\) as prediction of a specific class, y. For CNNs, we can approximate the highly non-linear function of y to be \(y \approx w^{\mathbf {T}}x + b\) where w and b are the weight and bias of the model. So ranking the influence of each pixel, p, can be posed as ranking the magnitude of the specific weight, \(w_p\), that influences the output y. The weight can be obtained as \(w_p = \frac{\partial y}{\partial x_p}\), which can be found by back-propagation.
However, unlike [13], where the input x is a 3 channel RGB image (\(z = 3\)), our input consists of 9 channels (\(z = 9\)), each a greyscale image of an MRI slice in the disc volume. Furthermore, instead of producing a 1 channel saliency map calculated from the maximum magnitude of w, \(\mathcal M = max_{z}|w |\) as in [13], the saliency map is also 9 dimensional, such that \(\mathcal M_z = |w_z |\) where \(z \in \{1 \dots 9\}\) since each input channel corresponds to an actual volumetric slice of the disc. Also, the final heat-map is computed from the average of multiple saliency maps produced from randomly augmented images using our training augmentation scheme; the resulting saliency map is transformed back to the original image space with the reverse of the augmentation. We find that aggressive augmentations is key to producing better localized hotspots. We visualize the evidence hotspots for some of the tasks on randomly selected discs in our test set in Fig. 3. The main visual difference from our maps to the maps shown in [13] is that our salient regions are more localized and more specific to the area that is the cause for the classification. We suspect that this might be because our classification tasks are more fine-grained, and because our input images are visually very similar.
Evidence Hotspots of the Test Set: Examples of disc volumes (upper in each pair) and their corresponding evidence hotspots (lower in each pair). The leftmost and rightmost images are the second and eighth slice for each disc, out of the full volume of 9 slices. Going from top to bottom are examples for each of the binary tasks: (i) upper endplate defects, (ii) lower endplate defects, (iii) upper marrow change, and (iv) lower marrow change. Pathological examples are shown for each radiological score/classification task, with endplate defects appearing as protrusions of the discs into the vertebral bodies, and marrow changes appearing as localized discolourations of the vertebral bodies near the vertebral endplates. Note that these hotspots localize extremely well to the assigned tasks e.g. in the lower endplate defects example the hotspots appear only in the lower endplate even though there are defects on the upper endplate. These examples are randomly selected on different patients. Further examples are given in the supplementary material.
4 Implementation Details
CNN architecture: The base network architecture is a modified version of the 1024 variant of the VGG-M network introduced in [2]. To maintain the aspect ratio of the discs, the input dimension is changed to \(112 \times 224 \times 9\) where 9 refers to the number of slices in a disc volume extracted from the full sagittal MR scan. Consequently, the Conv1 filters of VGG-M are changed to be \(7 \times 7 \times 9\) filters instead of the standard \(7 \times 7 \times 3\) filters for RGB images. We also omit the use of local response normalization after the Conv1 and Conv2 layers. The point at which to branch out the learning of the separate tasks are pre-selected and fixed prior to training. The layers succeeding the branch point are identical for each task except for the final FC8 layer, which has specific output dimensionality according to the number of classes in each task e.g. 5-way softmax for Pfirrmann, and 4-way softmax for disc narrowing classifications. Figure 2 shows a network with a branch point after Conv5. The networks were trained via the MatConvNet toolbox [14] using an NVIDIA Titan X GPU.
Disc volume extraction: The disc volumes are detected using the approach of [5, 7]. First a tight bounding volume is obtained about each of the seven vertebral bodies adjacent to the six radiologically labelled discs via the detection and regression steps. Then, from each pair of vertebral bodies, rough estimates of disc bounding volumes are obtained. Finally, the disc volumes for classification are defined as follows: the region is rotated within the sagittal slice so that the disc is horizontal and centered. The regions are resized to be the same dimension \(112 \times 224\) per slice; this 1:2 ratio is to ensure that the disc region would not include the upper and lower endplates of the adjacent vertebral bodies. Roughly \(40\,\%\) of each vertebrae, upper and lower, appear in each region of interest. The discs are aligned according to their mid-sagittal slices, and narrow discs with less than 9 slices are zero-padded slice-wise. Each slice of the disc is normalized such that the median intensity of its pair of vertebral bodies is 0.5 to mitigate against bias field effects. The range of the intensity inside the disc volume is set to be between 0 and 1 with higher intensity values clipped to 1.
Training: Training of the six tasks is done end-to-end simultaneously via stochastic gradient descent with momentum from scratch without any pre-training. The inputs are normalized with per-channel mean subtraction. The hyperparameters are: mini-batch size 256; momentum 0.9; weight decay 0.0005; initial learning rate 0.001, which is lowered by a factor of 10 as the error plateaus. The weights are initialized as [4] and normally reach convergence in about 1000 epochs.
Data augmentation details: We employ several aggressive random on-the-fly augmentation strategies during training. We include the methods suggested by [6] for natural images, and also define additional augmentations that are suited to sagittal scans. The training augmentation strategies are: (i) rotation with \(\theta \) = \(-15^{\circ }\) to \(15^{\circ }\), (ii) translation of \(\pm 32\) pixels in the x-axis, \(\pm 24\) pixels in the y-axis, \(\pm 2\) slices in the z-axis, (iii) rescaling with a scaling factor between \(90\,\%\) to \(110\,\%\), (iv) intensity variation between \(-0.1\) to 0.1, and (v) random slice-wise flip i.e. reflection of the slices across the mid-sagittal. At test time, the final prediction is calculated from the average of 54 predictions: (i) 8 patches, \(\pm 16\) pixels from the origin, alongside the centre patch, (ii) their slice-wise reflections, and (iii) sliding the slice window \(\pm 1\) across the volume.
5 Experiments and Results
Dataset and Radiological Gradings: The dataset is sourced from various centres with different MRI machines and protocols. Because of this, slice thickness also varies substantially from one scan to the other, ranging from 2.6 mm to 6.0 mm, with a median of 4 mm. The scans are T2 sagittal. The scans were annotated with various radiological scores (global, the whole spine, and local, per disc) by a single expert radiologist. In all, the dataset consists of 2009 patients from which we obtained 12018 individual discs, six discs per patient, and their labels. Some scans contain fewer than six discs but the majority show the complete lumbar region. We use a 80:10:10 split, on a per patient basis (1602:204:203), of the dataset to train, validate, and test the performance of our CNN model. The distribution of labels per disc for each classification task can be seen in Table 1.
Evaluation protocols: To evaluate our classification performance, we use average per-class accuracy which is suitable for highly imbalanced classifications. For comparison, we provide the average per-class intra-rater agreement which was calculated from two separate sets of labels by the same radiologist on a subset of the dataset which consists of 121 patients (726 discs), compared to the test set which consists of 203 patients (1217 discs). The intra-rater agreement serves as a good benchmark of performance since we are essentially limited to the quality of the label i.e. we can only be as good as the radiologist.
Model components and training: We investigate variations of the architecture by changing the branch point. Layers immediately after a branch point are duplicated for each of the six tasks e.g. for a network with a branch point after Conv4 we have six unique Conv5, FC6, FC7, and FC8 layers, one for each task. The accuracy for each task and the intra-rater agreement is given in Table 2. It can be seen that branching immediately after Conv5 is the best choice, and we use this configuration in subsequent comparisons. To obtain a standard deviation over the results, two models are trained for each experiment by swapping the validation and test sets. We also experimented with turning off data augmentation during both training and at test time, and found that there is a consistent decrease of 0.5 % in performance if test time augmentation is turned off, and the network overfits to the training set when augmentation is turned off during training. Overall, we achieve near-human performance, comparing our results to the intra-rater agreement, for all the learned tasks and consistently better results if the learned tasks are jointly learned. We also conducted further experiments on the effectiveness of multi-tasking which can be found in the supplementary material at http://www.robots.ox.ac.uk/~vgg/research/spine.
Comparison to the state-of-the-art: We evaluate our performance on Pfirrman grading and disc narrowing classifications using the test set and evaluation protocol of Lootus et al. [7]. We surpass their performance by +8.7 % (\(\mathbf 87.4\,\% \rightarrow \mathbf 96.1\,\% \)) for Pfirrmann grading, and +4.1 % (\(\mathbf 83.7\,\% \rightarrow \mathbf 87.8\,\% \)) for disc narrowing. It is important to note that in [7]: Pfirrmann grading is measured in terms of accuracy to \(\pm 1\) of the radiologist grade, and disc narrowing grading is simplified to a binary classification of normal/abnormal discs.
6 Summary and Future Work
We have shown that radiological scores and pathology hotspots can be predicted to an excellent standard using only the “weak” supervision of class labels. The proposed method is quite general, and although we have implemented it here for sagittal T2 scans, it could easily be applied to T1 scans, or indeed to any medical task where weak supervision is available.
References
Bar, Y., Diamant, I., Wolf, L., Lieberman, S., Konen, E., Greenspan, H.: Chest pathology detection using deep learning with non-medical training. In: ISBI (2015)
Chatfield, K., Simonyan, K., Vedaldi, A., Zisserman, A.: Return of the devil in the details: delving deep into convolutional nets. In: Proceedings of BMVC (2014)
Ghosh, S., Alomari, R.S., Chaudhary, V., Dhillon, G.: Computer-aided diagnosis for lumbar mri using heterogeneous classifiers. In: ISBI (2011)
Glorot, X., Bengio, Y.: Understanding the difficulty of training deep feedforward neural networks. In: Proceedings of AISTATS (2010)
Jamaludin, A., Kadir, T., Zisserman, A.: Automatic modic changes classication in spinal mri. In: MICCAI Workshop: CSI (2015)
Krizhevsky, A., Sutskever, I., Hinton, G.E.: ImageNet classification with deep convolutional neural networks. In: NIPS (2012)
Lootus, M.: Automated radiological analysis of Spinal MRI. Ph.D. thesis, University of Oxford (2015)
Oquab, M., Bottou, L., Laptev, I., Sivic, J.: Is object localization for free? weakly-supervised learning with convolutional neural networks. In: Proceedings of CVPR (2015)
Roberts, M.G., Pacheco, E.M., Mohankumar, R., Cootes, T.F., Adams, J.E.: Detection of vertebral fractures in DXA VFA images using statistical models of appearance and a semi-automatic segmentation. Osteoporos Int. 21(12), 2037–2046 (2010)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Heidelberg (2015). doi:10.1007/978-3-319-24574-4_28
Roth, H.R., Yao, J., Lu, L., Stieger, J., Burns, J.E., Summers, R.M.: Detection of sclerotic spine metastases viarandom aggregation of deep convolutionalneural network classifications. In: MICCAI Workshop: CSI (2014)
Shin, H.C., Roth, H.R., Gao, M., Lu, L., Xu, Z., Nogues, I., Yao, J., Mollura, D., Summers, R.M.: Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans. Med. Imaging 35(5), 1285–1298 (2016)
Simonyan, K., Vedaldi, A., Zisserman, A.: Deep inside convolutional networks: visualising image classification models and saliency maps. In: Workshop at International Conference on Learning Representations (2014)
Vedaldi, A., Lenc, K.: MatConvNet - convolutional neural networks for MATLAB. CoRR abs/1412.4564 (2014)
Acknowledgments
We are grateful for discussions with Prof. Jeremy Fairbank and Dr. Jill Urban, and Prof. Iain McCall for the radiological scores. This work was supported by the RCUK CDT in Healthcare Innovation (EP/G036861/1) and the EPSRC Programme Grant Seebibyte (EP/M013774/1). The data was obtained during the EC FP7 project (HEALTH-F2-2008-201626).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing AG
About this paper
Cite this paper
Jamaludin, A., Kadir, T., Zisserman, A. (2016). SpineNet: Automatically Pinpointing Classification Evidence in Spinal MRIs. In: Ourselin, S., Joskowicz, L., Sabuncu, M., Unal, G., Wells, W. (eds) Medical Image Computing and Computer-Assisted Intervention – MICCAI 2016. MICCAI 2016. Lecture Notes in Computer Science(), vol 9901. Springer, Cham. https://doi.org/10.1007/978-3-319-46723-8_20
Download citation
DOI: https://doi.org/10.1007/978-3-319-46723-8_20
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-46722-1
Online ISBN: 978-3-319-46723-8
eBook Packages: Computer ScienceComputer Science (R0)