Summary
RNA secondary structures are computed from primary sequences by means of a folding algorithm which uses a minimum free energy criterion. Free energies as well as replication and degradation rate constants are derived from secondary structures. These properties can be understood as highly sophisticated functions of the individual sequences whose values are mediated by the secondary structures. Such functions induce complex value landscapes on the space of sequences. The landscapes are analysed by random walk techniques, in particular autocorrelation functions and correlation lengths are computed. Free energy landscapes were found to be of AR(1) type. The rate constant landscapes, however, turned out to be more complex. In addition, gradient and adaptive walks are performed in order to get more insight into the complex structure of the landscapes.
Zusammenfassung
RNA-Sekundärstrukturen werden aus den Primärsequenzen mit Hilfe eines Computeralgorithmus berechnet, welcher einem Kriterium minimaler freier Energien folgt. Freie Energien, Replikations- oder Abbaugeschwindigkeitskonstanten werden aus den Sekundärstrukturen berechnet. Man kann daher diese Eigenschaften als komplizierte Funktionen der Sequenzen auffassen, deren Zahlenwerte durch Vermittlung der Sekundärstrukturen erhalten werden. Diese Funktionen induzieren hochkomplexe Bewertungslandschaften im Raum der Sequenzen. Die Landschaften werden mit Hilfe von Irrflugtechniken analysiert. Im einzelnen werden Autokorrelationsfunktionen und Korrelationslängen berechnet. Die freien Energie-Landschaften sind vom AR(1) Typ. Die von den Reaktionsgeschwindigkeitskonstanten abgeleiteten Landschaften stellten sich hingegen als komplexer heraus. Zusätzlich werden die Bewertungslandschaften auch noch mit Hilfe vonGradient undAdaptive Walks untersucht, um mehr Einblick in ihre komplexe Struktur zu gewinnen.
Similar content being viewed by others
References
Horwitz M. S. Z., Dube D. K., Loeb L. A. (1989) Genome31: 112
Joyce G. F. (1989) Gene82: 83
Tuerk C., Gold L. (1990) Science249: 505
Ellington A. D., Szostak J. W. (1990) Nature346: 818
Husimi Y., Keweloh C. (1987) Rev. Sci. Instrum.58: 1109
Biebricher C. K. (1988) Cold Spring Harbor Symp. Quant. Biol.52: 299
Spiegelman S. (1971) Quart. Rev. Biophys.4: 213
Eigen M. (1986) Chemica Scripta26B: 13
Kauffman S. A. (1986) J. Theor. Biol.119: 1
Lerner R. A., Tramontano A. (1988) Sci. Am.258/3: 42
Schulz P., Lerner R. A. (in press) At the cross-roads of chemistry and immunology: Catalytic antibodies. Science
Wright S. (1932) Proceedings of the Sixth International Congress on Genetics1: 356
Kauffman S. A., Levin S. (1987) J. theor. Biol.128: 11
Kauffman S. A. (1989) Adaptation on rugged fitness landscapes. In: Stein D. (ed.) Complex Systems (SFI Studies in the Science of Complexity). Addison-Wesley Longman, Redwood City, CA, pp. 527–618
Macken C. A., Perelson A. S. (1989) Proc. Natl. Acad. Sci. USA86: 6191
Fontana W., Schuster P. (1987) Biophys. Chem.26: 123
Fontana W., Schnabl W., Schuster P. (1989) Phys. Rev. A40: 3301
Schuster P. (1991) Complex optimization in an artificial RNA world. In: Farmer D., Langton C., Rasmussen S., Taylor C. (eds.) Artificial Life II (SFI Studies in the Sciences of Complexity, Vol. XII). Addison-Wesley Longman, Redwood City, CA
Eigen M., McCaskill J., Schuster P. (1988) J. Phys. Chem.92: 6881
Schuster P., Swetina J. (1988) Bull. Math. Biol.50: 635
Eigen M., McCaskill J., Schuster P. (1989) Adv. Chem. Phys.75: 149
Biebricher C. K., Eigen M., Gardiner jr., W. A. (in press) Quantitative analysis of selection and mutation in self-replicating RNA. In: Peliti L. (ed.) Biologically Inspired Physics (NATO Advanced Study Series)
Jaeger J. A., Turner D. H., Zuker M. (1989) Proc. Natl. Acad. Sci. USA86: 7706
Fontana W., Konings D. A. M., Schuster P. (1991) Statistics of RNA Secondary Structures (Preprint)
Sankoff D., Morin A.-M., Cedergren R. J. (1978) Can. J. Biochem.56: 440
Cech T. R. (1988) Gene73: 259
Le S.-Y., Zuker M. (1990) J. Mol. Biol.216: 729
Zuker M., Sankoff D. (1984) Bull. Math. Biol.46: 591
Zuker M. (1989) Science244: 48
Jaeger J. A., Turner D. H., Zuker M. (1990) Methods in Enzymology183: 281
McCaskill J. S. (1990) Biopolymers29: 1105
Hamming R. W. (1989) Coding and Information Theory, 2nd Ed. Prentice-Hall, Englewood Cliffs, NJ, pp. 44–47
Maynard Smith J. (1970) Nature225: 563
Shapiro B. A. (1988) CABIOS4: 387
Shapiro B. A., Zhang K. (1990) CABIOS6: 309
Sankoff D., Kruskal J. B. (1983) Time Warps, String Edits, and Macromolecules: The Theory and Practice of Sequence Comparison. Addison-Wesley, Reading
Tai K.-C. (1979) J. Ass. Computing Machinery26: 422
Hogeweg P., Hesper B. (1984) Nucleic Acids Research12: 67
Konings D. A. M. (1989) Pattern Analysis of RNA Secondary Structure (Proefschrift) Rijksuniversiteit te Utrecht
Konings D. A. M., Hogeweg P. (1989) J. Mol. Biol.207: 597
Fontana W., Konings D. A. M., Stadler P. F., Schuster P. (1991) Quantitative comparison and Statistics of RNA Secondary Structures (Preprint)
Karlin S., Taylor H. M. (1975) A First Course in Stochastic Processes, 2nd Ed. Academic Press, New York, pp. 455–461
Weinberger E. D. (1990) Biol. Cybern.63: 325
Sherrington D., Kirkpatrick S. (1975) Phys. Rev. Lettes35: 1792
Stadler P. F., Schnabl W. (1991) The Landscape of the Traveling Salesman Problem (Preprint)
Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilkson T., Turner D. H. (1986) Proc. Natl. Acad. Sci. USA83: 9373
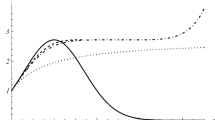
Fontana W., Stadler P. F., Griesmacher T., Weinberger E. D., Schuster P. (1991) Statistical Properties of RNA Free Energy Landscapes. A Study by Random Walk Techniques (Preprint)
Author information
Authors and Affiliations
Additional information
Dedicated to Prof. Dr. Dr. h.c. mult. Viktor Gutmann
Rights and permissions
About this article
Cite this article
Fontana, W., Griesmacher, T., Schnabl, W. et al. Statistics of landscapes based on free energies, replication and degradation rate constants of RNA secondary structures. Monatsh Chem 122, 795–819 (1991). https://doi.org/10.1007/BF00815919
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00815919