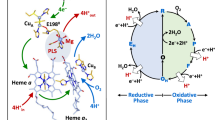
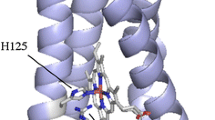
Abstract
Cytochrome-c (cyt-c), a multi-functional protein, plays a significant role in the electron transport chain, and thus is indispensable in the energy-production process. Besides being an important component in apoptosis, it detoxifies reactive oxygen species. Two hundred and eighty-five complete amino acid sequences of cyt-c from different species are known. Sequence analysis suggests that the number of amino acid residues in most mitochondrial cyts-c is in the range 104 ± 10, and amino acid residues at only few positions are highly conserved throughout evolution. These highly conserved residues are Cys14, Cys17, His18, Gly29, Pro30, Gly41, Asn52, Trp59, Tyr67, Leu68, Pro71, Pro76, Thr78, Met80, and Phe82. These are also known as “key residues”, which contribute significantly to the structure, function, folding, and stability of cyt-c. The three-dimensional structure of cyt-c from ten eukaryotic species have been determined using X-ray diffraction studies. Structure analysis suggests that the tertiary structure of cyt-c is almost preserved along the evolutionary scale. Furthermore, residues of N/C-terminal helices Gly6, Phe10, Leu94, and Tyr97 interact with each other in a specific manner, forming an evolutionary conserved interface. To understand the role of evolutionary conserved residues on structure, stability, and function, numerous studies have been performed in which these residues were substituted with different amino acids. In these studies, structure deals with the effect of mutation on secondary and tertiary structure measured by spectroscopic techniques; stability deals with the effect of mutation on T m (midpoint of heat denaturation), ∆G D (Gibbs free energy change on denaturation) and folding; and function deals with the effect of mutation on electron transport, apoptosis, cell growth, and protein expression. In this review, we have compiled all these studies at one place. This compilation will be useful to biochemists and biophysicists interested in understanding the importance of conservation of certain residues throughout the evolution in preserving the structure, function, and stability in proteins.



Similar content being viewed by others
References
Pettigrew GW, Moore GR (1987) Cytochrome c: biological aspects. Springer, Berlin Heidelberg New York
Moore GR, Pettigrew GW (1990) Cytochromes c: evolutionary, structural and physicochemical aspects. Springer, Berlin Heidelberg New York, pp 831–833
Kalanxhi E, Wallace CJ (2007) Cytochrome c impaled: investigation of the extended lipid anchorage of a soluble protein to mitochondrial membrane models. Biochem J 407:179–187
Orrenius S (2004) Mitochondrial regulation of apoptotic cell death. Toxicol Lett 149:19–23
Abdullaev Z, Bodrova ME, Chernyak BV et al (2002) A cytochrome c mutant with high electron transfer and antioxidant activities but devoid of apoptogenic effect. Biochem J 362:749–754
Pereverzev MO, Vygodina TV, Konstantinov AA, Skulachev VP (2003) Cytochrome c, an ideal antioxidant. Biochem Soc Trans 31:1312–1315
Skulachev VP (1998) Cytochrome c in the apoptotic and antioxidant cascades. FEBS Lett 423:275–280
Belikova NA, Vladimirov YA, Osipov AN et al (2006) Peroxidase activity and structural transitions of cytochrome c bound to cardiolipin-containing membranes. Biochemistry 45:4998–5009
Garcia-Heredia JM, Diaz-Moreno I, Nieto PM et al (2010) Nitration of tyrosine 74 prevents human cytochrome c to play a key role in apoptosis signaling by blocking caspase-9 activation. Biochim Biophys Acta 1797:981–993
Kapralov AA, Kurnikov IV, Vlasova II et al (2007) The hierarchy of structural transitions induced in cytochrome c by anionic phospholipids determines its peroxidase activation and selective peroxidation during apoptosis in cells. Biochemistry 46:14232–14244
Kapralov AA, Yanamala N, Tyurina YY et al (2011) Topography of tyrosine residues and their involvement in peroxidation of polyunsaturated cardiolipin in cytochrome c/cardiolipin peroxidase complexes. Biochim Biophys Acta 1808:2147–2155
Pecina P, Borisenko GG, Belikova NA et al (2010) Phosphomimetic substitution of cytochrome C tyrosine 48 decreases respiration and binding to cardiolipin and abolishes ability to trigger downstream caspase activation. Biochemistry 49:6705–6714
Ying T, Wang ZH, Lin YW et al (2009) Tyrosine-67 in cytochrome c is a possible apoptotic trigger controlled by hydrogen bonds via a conformational transition. Chem Commun (Camb) 30:4512–4514
Liu Z, Lin H, Ye S et al (2006) Remarkably high activities of testicular cytochrome c in destroying reactive oxygen species and in triggering apoptosis. Proc Natl Acad Sci USA 103:8965–8970
Koonin EV, Aravind L (2002) Origin and evolution of eukaryotic apoptosis: the bacterial connection. Cell Death Differ 9:394–404
Tezcan FA, Findley WM, Crane BR et al (2002) Using deeply trapped intermediates to map the cytochrome c folding landscape. Proc Natl Acad Sci USA 99:8626–8630
Yeh SR, Rousseau DL (1998) Folding intermediates in cytochrome c. Nat Struct Biol 5:222–228
Cianetti S, Negrerie M, Vos MH et al (2004) Photodissociation of heme distal methionine in ferrous cytochrome C revealed by subpicosecond time-resolved resonance Raman spectroscopy. J Am Chem Soc 126:13932–13933
Banci L, Bertini I, Rosato A, Varani G (1999) Mitochondrial cytochromes c: a comparative analysis. J Biol Inorg Chem 4:824–837
Dickerson RE (1971) Sequence and structure homologies in bacterial and mammalian-type cytochromes. J Mol Biol 57:1–15
Ferguson-Miller S, Brautigan DL, Margoliash E (1976) Correlation of the kinetics of electron transfer activity of various eukaryotic cytochromes c with binding to mitochondrial cytochrome c oxidase. J Biol Chem 251:1104–1115
Filosa A, English AM (2000) Probing local thermal stabilities of bovine, horse, and tuna ferricytochromes c at pH 7. J Biol Inorg Chem 5:448–454
Fredericks ZL, Pielak GJ (1993) Exploring the interface between the N- and C-terminal helices of cytochrome c by random mutagenesis within the C-terminal helix. Biochemistry 32:929–936
Knapp JA, Pace CN (1974) Guanidine hydrochloride and acid denaturation of horse, cow, and Candida krusei cytochromes c. Biochemistry 13:1289–1294
McLendon G, Smith M (1978) Equilibrium and kinetic studies of unfolding of homologous cytochromes c. J Biol Chem 253:4004–4008
Yuan X, Hawkridge FM, Chlebowski JF (1993) Thermodynamic and kinetic studies of cytochrome c from different species. J Electroanal Chem 350:29–42
Moza B, Qureshi SH, Ahmad F (2003) Equilibrium studies of the effect of difference in sequence homology on the mechanism of denaturation of bovine and horse cytochromes-c. Biochim Biophys Acta 1646:49–56
Hampsey DM, Das G, Sherman F (1986) Amino acid replacements in yeast iso-1-cytochrome c. Comparison with the phylogenetic series and the tertiary structure of related cytochromes c. J Biol Chem 261:3259–3271
Smith M (1986) Site-directed mutagenesis. Phil Trans R SOC Lond A 317:295–304
Zoller MJ, Smith M (1983) Oligonucleotide-directed mutagenesis of DNA fragments cloned into M13 vectors. Methods Enzymol 100:468–500
Sherman F, Stewart JW (1974) Variation of mutagenic action on nonsense mutants at different sites in the iso-1-cytochrome c gene of yeast. Genetics 78:97–113
Sherman F, Stewart JW, Parker JH et al (1968) The mutational alteration of the primary structure of yeast iso-1-cytochrome c. J Biol Chem 243:5446–5456
Brown GC, Borutaite V (2008) Regulation of apoptosis by the redox state of cytochrome c. Biochim Biophys Acta 1777:877–881
Jemmerson R, Liu J, Hausauer D et al (1999) A conformational change in cytochrome c of apoptotic and necrotic cells is detected by monoclonal antibody binding and mimicked by association of the native antigen with synthetic phospholipid vesicles. Biochemistry 38:3599–3609
Ow YP, Green DR, Hao Z, Mak TW (2008) Cytochrome c: functions beyond respiration. Nat Rev Mol Cell Biol 9:532–542
Huttemann M, Pecina P, Rainbolt M et al (2011) The multiple functions of cytochrome c and their regulation in life and death decisions of the mammalian cell: from respiration to apoptosis. Mitochondrion 11:369–381
Rumbley JN, Hoang L, Englander SW (2002) Recombinant equine cytochrome c in Escherichia coli: high-level expression, characterization, and folding and assembly mutants. Biochemistry 41:13894–13901
Russell BS, Melenkivitz R, Bren KL (2000) NMR investigation of ferricytochrome c unfolding: detection of an equilibrium unfolding intermediate and residual structure in the denatured state. Proc Natl Acad Sci USA 97:8312–8317
Sambongi Y, Uchiyama S, Kobayashi Y et al (2002) Cytochrome c from a thermophilic bacterium has provided insights into the mechanisms of protein maturation, folding, and stability. Eur J Biochem 269:3355–3361
Santucci R, Bongiovanni C, Mei G et al (2000) Anion size modulates the structure of the A state of cytochrome c. Biochemistry 39:12632–12638
Sauder JM, Roder H (1998) Amide protection in an early folding intermediate of cytochrome c. Fold Des 3:293–301
Schweitzer-Stenner R, Shah R, Hagarman A, Dragomir I (2007) Conformational substates of horse heart cytochrome c exhibit different thermal unfolding of the heme cavity. J Phys Chem B 111:9603–9607
Smith LJ, Kahraman A, Thornton JM (2010) Heme proteins–diversity in structural characteristics, function, and folding. Proteins 78:2349–2368
Telford JR, Tezcan FA, Gray HB, Winkler JR (1999) Role of ligand substitution in ferrocytochrome c folding. Biochemistry 38:1944–1949
Thomas YG, Goldbeck RA, Kliger DS (2000) Characterization of equilibrium intermediates in denaturant-induced unfolding of ferrous and ferric cytochromes c using magnetic circular dichroism, circular dichroism, and optical absorption spectroscopies. Biopolymers 57:29–36
Zhong S, Rousseau DL, Yeh SR (2004) Modulation of the folding energy landscape of cytochrome c with salt. J Am Chem Soc 126:13934–13935
Goldbeck RA, Chen E, Kliger DS (2009) Early events, kinetic intermediates and the mechanism of protein folding in cytochrome c. Int J Mol Sci 10:1476–1499
Levinthal C (1968) Are there pathways for protein folding? J Chim Phys 65:44–45
Dill KA, Phillips AT, Rosen JB (1997) Protein structure and energy landscape dependence on sequence using a continuous energy function. J Comput Biol 4:227–239
Dill KA, Chan HS (1997) From Levinthal to pathways to funnels. Nat Struct Biol 4:10–19
Dill KA, Ozkan SB, Weikl TR et al (2007) The protein folding problem: when will it be solved? Curr Opin Struct Biol 17:342–346
Dill KA, Ozkan SB, Shell MS, Weikl TR (2008) The protein folding problem. Annu Rev Biophys 37:289–316
Jha SK, Udgaonkar JB (2010) Free energy barriers in protein folding and unfolding reactions. Curr Sci 99:457–475
Sinha KK, Udgaonkar JB (2009) Early events in protein folding. Curr Sci 96:1053–1070
Bolen DW, Rose GD (2008) Structure and energetics of the hydrogen-bonded backbone in protein folding. Annu Rev Biochem 77:339–362
Daggett V, Fersht AR (2003) Is there a unifying mechanism for protein folding? Trends Biochem Sci 28:18–25
Vertrees J, Wrabl JO, Hilser VJ (2009) Energetic profiling of protein folds. Methods Enzymol 455:299–327
Arai M, Kuwajima K (2000) Role of the molten globule state in protein folding. Adv Protein Chem 53:209–282
Ptitsyn OB (1995) Structures of folding intermediates. Curr Opin Struct Biol 5:74–78
Ptitsyn OB (1995) How the molten globule became. Trends Biochem Sci 20:376–379
Udgaonkar JB (2008) Multiple routes and structural heterogeneity in protein folding. Annu Rev Biophys 37:489–510
Yamada S, Bouley Ford ND, Keller GE et al (2013) Snapshots of a protein folding intermediate. Proc Natl Acad Sci USA 110:1606–1610
Travaglini-Allocatelli C, Gianni S, Brunori M (2004) A common folding mechanism in the cytochrome c family. Trends Biochem Sci 29:535–541
Thielges MC, Zimmermann J, Dawson PE, Romesberg FE (2009) The determinants of stability and folding in evolutionarily diverged cytochromes c. J Mol Biol 388:159–167
Qureshi SH, Moza B, Yadav S, Ahmad F (2003) Conformational and thermodynamic characterization of the molten globule state occurring during unfolding of cytochromes-c by weak salt denaturants. Biochemistry 42:1684–1695
Ahmad Z, Ahmad F (1994) Physico-chemical characterization of products of unfolding of cytochrome c by calcium chloride. Biochim Biophys Acta 1207:223–230
Alam Khan MK, Rahaman MH, Hassan MI et al (2010) Conformational and thermodynamic characterization of the premolten globule state occurring during unfolding of the molten globule state of cytochrome c. J Biol Inorg Chem 15:1319–1329
Bai Y (1999) Kinetic evidence for an on-pathway intermediate in the folding of cytochrome c. Proc Natl Acad Sci USA 96:477–480
Bhuyan AK, Udgaonkar JB (2001) Folding of horse cytochrome c in the reduced state. J Mol Biol 312:1135–1160
Bhuyan AK, Rao DK, Prabhu NP (2005) Protein folding in classical perspective: folding of horse cytochrome c. Biochemistry 44:3034–3040
Colon W, Roder H (1996) Kinetic intermediates in the formation of the cytochrome c molten globule. Nat Struct Biol 3:1019–1025
Englander SW (2000) Protein folding intermediates and pathways studied by hydrogen exchange. Annu Rev Biophys Biomol Struct 29:213–238
Goto Y, Hagihara Y, Hamada D et al (1993) Acid-induced unfolding and refolding transitions of cytochrome c: a three-state mechanism in H2O and D2O. Biochemistry 32:11878–11885
Hamada D, Hoshino M, Kataoka M et al (1993) Intermediate conformational states of apocytochrome c. Biochemistry 32:10351–10358
Prabhu NP, Kumar R, Bhuyan AK (2004) Folding barrier in horse cytochrome c: support for a classical folding pathway. J Mol Biol 337:195–208
Moza B, Qureshi SH, Islam A et al (2006) A unique molten globule state occurs during unfolding of cytochrome c by LiClO4 near physiological pH and temperature: structural and thermodynamic characterization. Biochemistry 45:4695–4702
Lyubovitsky JG, Gray HB, Winkler JR (2002) Mapping the cytochrome C folding landscape. J Am Chem Soc 124:5481–5485
Pletneva EV, Gray HB, Winkler JR (2005) Many faces of the unfolded state: conformational heterogeneity in denatured yeast cytochrome C. J Mol Biol 345:855–867
Pletneva EV, Gray HB, Winkler JR (2005) Snapshots of cytochrome c folding. Proc Natl Acad Sci USA 102:18397–18402
Pletneva EV, Zhao Z, Kimura T et al (2007) Probing the cytochrome c’ folding landscape. J Inorg Biochem 101:1768–1775
Bandi S, Bowler BE (2008) Probing the bottom of a folding funnel using conformationally gated electron transfer reactions. J Am Chem Soc 130:7540–7541
Hammack B, Godbole S, Bowler BE (1998) Cytochrome c folding traps are not due solely to histidine-heme ligation: direct demonstration of a role for N-terminal amino group-heme ligation. J Mol Biol 275:719–724
Marmorino JL, Lehti M, Pielak GJ (1998) Native tertiary structure in an A-state. J Mol Biol 275:379–388
Gianni S, Travaglini-Allocatelli C, Cutruzzola F et al (2001) Snapshots of protein folding. A study on the multiple transition state pathway of cytochrome c(551) from Pseudomonas aeruginosa. J Mol Biol 309:1177–1187
Gianni S, Travaglini-Allocatelli C, Cutruzzola F et al (2003) Parallel pathways in cytochrome c(551) folding. J Mol Biol 330:1145–1152
Travaglini-Allocatelli C, Cutruzzola F, Bigotti MG et al (1999) Folding mechanism of Pseudomonas aeruginosa cytochrome c551: role of electrostatic interactions on the hydrophobic collapse and transition state properties. J Mol Biol 289:1459–1467
Travaglini-Allocatelli C, Gianni S, Morea V et al (2003) Exploring the cytochrome c folding mechanism: cytochrome c552 from thermus thermophilus folds through an on-pathway intermediate. J Biol Chem 278:41136–41140
Alam Khan MK, Das U, Rahaman MH et al (2009) A single mutation induces molten globule formation and a drastic destabilization of wild-type cytochrome c at pH 6.0. J Biol Inorg Chem 14:751–760
Bertini I, Turano P, Vasos PR et al (2004) Cytochrome c and SDS: a molten globule protein with altered axial ligation. J Mol Biol 336:489–496
Nakamura S, Seki Y, Katoh E, Kidokoro S (2011) Thermodynamic and structural properties of the acid molten globule state of horse cytochrome C. Biochemistry 50:3116–3126
Wittung-Stafshede P (1998) A stable, molten-globule-like cytochrome c. Biochim Biophys Acta 1382:324–332
Khan MK, Rahaman H, Ahmad F (2011) Conformation and thermodynamic stability of pre-molten and molten globule states of mammalian cytochromes-c. Metallomics 3:327–338
Alber T (1989) Mutational effects on protein stability. Annu Rev Biochem 58:765–798
Santucci R, Ascoli F (1997) The Soret circular dichroism spectrum as a probe for the heme Fe(III)-Met(80) axial bond in horse cytochrome c. J Inorg Biochem 68:211–214
Schejter A, Plotkin B, Vig I (1991) The reactivity of cytochrome c with soft ligands. FEBS Lett 280:199–201
Davis LA, Schejter A, Hess GP (1974) Alkaline isomerization of oxidized cytochrome c. Equilibrium and kinetic measurements. J Biol Chem 249:2624–2632
Ferrer JC, Guillemette JG, Bogumil R et al (1993) Identification of Lys79 as an iron ligand in one form of alkaline yeast iso-1-ferricytochrome c. J Am Chem Soc 90:7507
Feinberg BA, Petro L, Hock G et al (1999) Using entropies of reaction to predict changes in protein stability: tyrosine-67-phenylalanine variants of rat cytochrome c and yeast Iso-1 cytochromes c. J Pharm Biomed Anal 19:115–125
Battistuzzi G, Borsari M, Cowan JA et al (2002) Control of cytochrome C redox potential: axial ligation and protein environment effects. J Am Chem Soc 124:5315–5324
Kassner RJ (1973) A theoretical model for the effects of local nonpolar heme environments on the redox potentials in cytochromes. J Am Chem Soc 95:2674–2677
Stellwagen E (1978) Haem exposure as the determinate of oxidation-reduction potential of haem proteins. Nature 275:73–74
Bertrand P, Mbarki O, Asso M et al (1995) Control of the redox potential in c-type cytochromes: importance of the entropic contribution. Biochemistry 34:11071–11079
Churg AK, Warshel A (1986) Control of the redox potential of cytochrome c and microscopic dielectric effects in proteins. Biochemistry 25:1675–1681
Rees DC (1985) Electrostatic influence on energetics of electron transfer reactions. Proc Natl Acad Sci USA 82:3082–3085
Tai H, Mikami S, Irie K et al (2010) Role of a highly conserved electrostatic interaction on the surface of cytochrome C in control of the redox function. Biochemistry 49:42–48
Wells JA (1990) Additivity of mutational effects in proteins. Biochemistry 29:8509–8517
Hoang L, Maity H, Krishna MM et al (2003) Folding units govern the cytochrome c alkaline transition. J Mol Biol 331:37–43
Lett CM, Rosu-Myles MD, Frey HE, Guillemette JG (1999) Rational design of a more stable yeast iso-1-cytochrome c. Biochim Biophys Acta 1432:40–48
Sanishvili R, Volz KW, Westbrook EM, Margoliash E (1995) The low ionic strength crystal structure of horse cytochrome c at 2.1 Å resolution and comparison with its high ionic strength counterpart. Structure 3:707–716
Mirkin N, Jaconcic J, Stojanoff V, Moreno A (2008) High resolution X-ray crystallographic structure of bovine heart cytochrome c and its application to the design of an electron transfer biosensor. Proteins 70:83–92
Bushnell GW, Louie GV, Brayer GD (1990) High-resolution three-dimensional structure of horse heart cytochrome c. J Mol Biol 214:585–595
Louie GV, Brayer GD (1990) High-resolution refinement of yeast iso-1-cytochrome c and comparisons with other eukaryotic cytochromes c. J Mol Biol 214:527–555
Ptitsyn OB (1998) Protein folding and protein evolution: common folding nucleus in different subfamilies of c-type cytochromes? J Mol Biol 278:655–666
Fersht AR (1997) Nucleation mechanisms in protein folding. Curr Opin Struct Biol 7:3–9
Colon W, Elove GA, Wakem LP et al (1996) Side chain packing of the N- and C-terminal helices plays a critical role in the kinetics of cytochrome c folding. Biochemistry 35:5538–5549
Roder H, Colon W (1997) Kinetic role of early intermediates in protein folding. Curr Opin Struct Biol 7:15–28
Marmorino JL, Pielak GJ (1995) A native tertiary interaction stabilizes the a state of cytochrome c. Biochemistry 34:3140–3143
Matthews BW (1993) Structural and genetic analysis of protein stability. Annu Rev Biochem 62:139–160
Takeda T, Sonoyama T, Takayama SJ et al (2009) Correlation between the stability and redox potential of three homologous cytochromes c from two thermophiles and one mesophile. Biosci Biotechnol Biochem 73:366–371
Terui N, Tachiiri N, Matsuo H et al (2003) Relationship between redox function and protein stability of cytochromes c. J Am Chem Soc 125:13650–13651
Herbaud ML, Aubert C, Durand MC et al (2000) Escherichia coli is able to produce heterologous tetraheme cytochrome c(3) when the ccm genes are co-expressed. Biochim Biophys Acta 1481:18–24
Pettigrew GW, Leaver JL, Meyer TE, Ryle AP (1975) Purification, properties and amino acid sequence of atypical cytochrome c from two protozoa, Euglena gracilis and Crithidia oncopelti. Biochem J 147:291–302
Priest JW, Hajduk SL (1992) Cytochrome c reductase purified from Crithidia fasciculata contains an atypical cytochrome c1. J Biol Chem 267:20188–20195
Rios-Velazquez C, Cox RL, Donohue TJ (2001) Characterization of Rhodobacter sphaeroides cytochrome c(2) proteins with altered heme attachment sites. Arch Biochem Biophys 389:234–244
Barker PD, Ferguson SJ (1999) Still a puzzle: why is haem covalently attached in c-type cytochromes? Structure 7:R281–R290
Thony-Meyer L (2000) Haem-polypeptide interactions during cytochrome c maturation. Biochim Biophys Acta 1459:316–324
Stellwagen E, Cass R (1974) Alkaline isomerization of ferricytochrome C from Euglena gracilis. Biochem Biophys Res Commun 60:371–375
Bowman SE, Bren KL (2008) The chemistry and biochemistry of heme c: functional bases for covalent attachment. Nat Prod Rep 25:1118–1130
Hampsey DM, Das G, Sherman F (1988) Yeast iso-1-cytochrome c: genetic analysis of structural requirements. FEBS Lett 231:275–283
Rosell FI, Mauk AG (2002) Spectroscopic properties of a mitochondrial cytochrome C with a single thioether bond to the heme prosthetic group. Biochemistry 41:7811–7818
Hennig B, Neupert W (1983) Biogenesis of cytochrome c in Neurospora crassa. Methods Enzymol 97:261–274
Dumont MD, Mathews AJ, Nall BT et al (1990) Differential stability of two apo-isocytochromes c in the yeast Saccharomyces cerevisiae. J Biol Chem 265:2733–2739
Wang X, Dumont ME, Sherman F (1996) Sequence requirements for mitochondrial import of yeast cytochrome c. J Biol Chem 271:6594–6604
Mavridou DA, Stevens JM, Monkemeyer L et al (2012) A pivotal heme-transfer reaction intermediate in cytochrome c biogenesis. J Biol Chem 287:2342–2352
Cowley AB, Lukat-Rodgers GS, Rodgers KR, Benson DR (2004) A possible role for the covalent heme-protein linkage in cytochrome c revealed via comparison of N-acetylmicroperoxidase-8 and a synthetic, monohistidine-coordinated heme peptide. Biochemistry 43:1656–1666
Dumont ME, Corin AF, Campbell GA (1994) Noncovalent binding of heme induces a compact apocytochrome c structure. Biochemistry 33:7368–7378
Kang X, Carey J (1999) Role of heme in structural organization of cytochrome c probed by semisynthesis. Biochemistry 38:15944–15951
Lu Y, Casimiro DR, Bren KL et al (1993) Structurally engineered cytochromes with unusual ligand-binding properties: expression of Saccharomyces cerevisiae Met-80–> Ala iso-1-cytochrome c. Proc Natl Acad Sci USA 90:11456–11459
Raphael AL (1991) Semisynthesis of Axial-Ligand (position 80) mutants of cytochrome c. J Am Chem Soc 82:1038–1040
Rux JJ, Dawson JH (1991) Magnetic circular dichroism spectroscopy as a probe of axial heme ligand replacement in semisynthetic mutants of cytochrome c. FEBS Lett 290:49–51
Silkstone G, Jasaitis A, Wilson MT, Vos MH (2007) Ligand dynamics in an electron transfer protein. Picosecond geminate recombination of carbon monoxide to heme in mutant forms of cytochrome c. J Biol Chem 282:1638–1649
Silkstone G, Stanway G, Brzezinski P, Wilson MT (2002) Production and characterisation of Met80X mutants of yeast iso-1-cytochrome c: spectral, photochemical and binding studies on the ferrous derivatives. Biophys Chem 98:65–77
Bagel ova J, Gazova Z, Valusova E, Antalik M (2001) Conformational stability of ferricytochrome c near the heme in its complex with heparin in alkaline pH. Carbohydr Polym 135:980–986
Banci L, Bertini I, Bren KL et al (1995) Three-dimensional solution structure of the cyanide adduct of a Met80Ala variant of Saccharomyces cerevisiae iso-1-cytochrome c. Identification of ligand-residue interactions in the distal heme cavity. Biochemistry 34:11385–11398
Satoh T, Itoga A, Isogai Y et al (2002) Increasing the conformational stability by replacement of heme axial ligand in c-type cytochrome. FEBS Lett 531:543–547
Yeh SR, Takahashi S, Fan B, Rousseau DL (1997) Ligand exchange during cytochrome c folding. Nat Struct Biol 4:51–56
Elove GA, Bhuyan AK, Roder H (1994) Kinetic mechanism of cytochrome c folding: involvement of the heme and its ligands. Biochemistry 33:6925–6935
Hagen SJ, Latypov RF, Dolgikh DA, Roder H (2002) Rapid intrachain binding of histidine-26 and histidine-33 to heme in unfolded ferrocytochrome C. Biochemistry 41:1372–1380
Takano T, Dickerson RE (1981) Conformation change of cytochrome c. I. Ferrocytochrome c structure refined at 1.5 Å resolution. J Mol Biol 153:79–94
Dyson HJ, Beattie JK (1982) Spin state and unfolding equilibria of ferricytochrome c in acidic solutions. J Biol Chem 257:2267–2273
Greenwood C, Palmer G (1965) Evidence for the existence of two functionally distinct forms cytochrome c manomer at alkaline pH. J Biol Chem 240:3660–3663
Schejter A, George P (1964) The 695-nm band of ferricytochrome C and its relationship to protein conformation. Biochemistry 3:1045–1049
Wallace CJ, Clark-Lewis I (1992) Functional role of heme ligation in cytochrome c. Effects of replacement of methionine 80 with natural and non-natural residues by semisynthesis. J Biol Chem 267:3852–3861
George P, Glauser SC, Schejter A (1967) The reactivity of ferricytochrome c with ionic ligands. J Biol Chem 242:1690–1695
Babul J, Stellwagen E (1971) The existence of heme-protein coordinate-covalent bonds in denaturing solvents. Biopolymers 10:2359–2361
Muthukrishnan K, Nall BT (1991) Effective concentrations of amino acid side chains in an unfolded protein. Biochemistry 30:4706–4710
Stellwagen E, Rysavy R, Babul G (1972) The conformation of horse heart apocytochrome c. J Biol Chem 247:8074–8077
Tsong TY (1975) An acid induced conformational transition of denatured cytochrome c in urea and guanidine hydrochloride solutions. Biochemistry 14:1542–1547
Droghetti E, Oellerich S, Hildebrandt P, Smulevich G (2006) Heme coordination states of unfolded ferrous cytochrome C. Biophys J 91:3022–3031
Cutler RL, Pielak GJ, Mauk AG, Smith M (1987) Replacement of cysteine-107 of Saccharomyces cerevisiae iso-1-cytochrome c with threonine: improved stability of the mutant protein. Protein Eng 1:95–99
Allen JW, Ferguson SJ (2003) Variation of the axial haem ligands and haem-binding motif as a probe of the Escherichia coli c-type cytochrome maturation (Ccm) system. Biochem J 375:721–728
Allen JW, Leach N, Ferguson SJ (2005) The histidine of the c-type cytochrome CXXCH haem-binding motif is essential for haem attachment by the Escherichia coli cytochrome c maturation (Ccm) apparatus. Biochem J 389:587–592
Bowman SE, Bren KL (2010) Variation and analysis of second-sphere interactions and axial histidinate character in c-type cytochromes. Inorg Chem 49:7890–7897
Garcia-Rubio I, Braun M, Gromov I et al (2007) Axial coordination of heme in ferric CcmE chaperone characterized by EPR spectroscopy. Biophys J 92:1361–1373
Takahashi A, Kurahashi T, Fujii H (2009) Effect of imidazole and phenolate axial ligands on the electronic structure and reactivity of oxoiron(IV) porphyrin pi-cation radical complexes: drastic increase in oxo-transfer and hydrogen abstraction reactivities. Inorg Chem 48:2614–2625
Fumo G, Spitzer JS, Fetrow JS (1995) A method of directed random mutagenesis of the yeast chromosome shows that the iso-1-cytochrome c heme ligand His18 is essential. Gene 164:33–39
Yeh SR, Rousseau DL (1999) Ligand exchange during unfolding of cytochrome c. J Biol Chem 274:17853–17859
Casalini S, Battistuzzi G, Borsari M et al (2010) Electron transfer properties and hydrogen peroxide electrocatalysis of cytochrome c variants at positions 67 and 80. J Phys Chem B 114:1698–1706
Senn H, Wuthrich K (1985) Amino acid sequence, haem-iron co-ordination geometry and functional properties of mitochondrial and bacterial c-type cytochromes. Q Rev Biophys 18:111–134
Raphael AL, Gray HB (1989) Axial ligand replacement in horse heart cytochrome c by semisynthesis. Proteins 6:338–340
Battistuzzi G, Bortolotti CA, Bellei M et al (2012) Role of Met80 and Tyr67 in the low-pH conformational equilibria of cytochrome c. Biochemistry 51:5967–5978
Ferri T, Poscia A, Ascoli F, Santucci R (1996) Direct electrochemical evidence for an equilibrium intermediate in the guanidine-induced unfolding of cytochrome c. Biochim Biophys Acta 1298:102–108
Santucci R, Brunori M, Ascoli F (1987) Unfolding and flexibility in hemoproteins shown in the case of carboxymethylated cytochrome c. Biochim Biophys Acta 914:185–189
Santucci R, Giartosio A, Ascoli F (1989) Structural transitions of carboxymethylated cytochrome c: calorimetric and circular dichroic studies. Arch Biochem Biophys 275:496–504
Bren KL, Gray HB (1993) Structurally engineered cytochromes with novel ligand binding sites: oxy and carbon monoxy derivatives of semisynthetic horse heart Ala80 cytochrome c. J Am Chem Soc 115:10382
Brunori M, Wilson MT, Antonini E (1972) Properties of modified cytochromes. I. Equilibrium and kinetics of the pH-dependent transition in carboxymethylated horse heart cytochrome c. J Biol Chem 247:6076–6081
Wilson MT, Brunori M, Rotilio GC, Antonini E (1973) Properties of modified cytochromes. II. Ligand binding to reduced carboxymethyl cytochrome c. J Biol Chem 248:8162–8169
Flynn PF, Bieber Urbauer RJ, Zhang H et al (2001) Main chain and side chain dynamics of a heme protein: 15N and 2H NMR relaxation studies of R. capsulatus ferrocytochrome c2. Biochemistry 40:6559–6569
Casalini S, Battistuzzi G, Borsari M et al (2008) Electron transfer and electrocatalytic properties of the immobilized methionine80alanine cytochrome c variant. J Phys Chem B 112:1555–1563
Indiani C, de Sanctis G, Neri F et al (2000) Effect of pH on axial ligand coordination of cytochrome c” from Methylophilus methylotrophus and horse heart cytochrome c. Biochemistry 39:8234–8242
Mathews FS (1985) The structure, function and evolution of cytochromes. Prog Biophys Mol Biol 45:1–56
Auld DS, Young GB, Saunders AJ et al (1993) Probing weakly polar interactions in cytochrome c. Protein Sci 2:2187–2197
Gao Y, Boyd J, Williams RJ, Pielak GJ (1990) Assignment of proton resonances, identification of secondary structural elements, and analysis of backbone chemical shifts for the C102T variant of yeast iso-1-cytochrome c and horse cytochrome c. Biochemistry 29:6994–7003
Roder H, Elove GA, Englander SW (1988) Structural characterization of folding intermediates in cytochrome c by H-exchange labelling and proton NMR. Nature 335:700–704
Efimov AV (1984) A novel super-secondary structure of proteins and the relation between the structure and the amino acid sequence. FEBS Lett 166:33–38
Richardson JS, Richardson DC (1988) Helix lap-joints as ion-binding sites: DNA-binding motifs and Ca-binding “EF hands” are related by charge and sequence reversal. Proteins 4:229–239
Pielak GJ, Auld DS, Beasley JR et al (1995) Protein thermal denaturation, side-chain models, and evolution: amino acid substitutions at a conserved helix-helix interface. Biochemistry 34:3268–3276
Auld DS, Pielak GJ (1991) Constraints on amino acid substitutions in the N-terminal helix of cytochrome c explored by random mutagenesis. Biochemistry 30:8684–8690
Pelletier H, Kraut J (1992) Crystal structure of a complex between electron transfer partners, cytochrome c peroxidase and cytochrome c. Science 258:1748–1755
Sosnick TR, Mayne L, Hiller R, Englander SW (1994) The barriers in protein folding. Nat Struct Biol 1:149–156
Wu LC, Laub PB, Elove GA et al (1993) A noncovalent peptide complex as a model for an early folding intermediate of cytochrome c. Biochemistry 32:10271–10276
Berghuis AM, Brayer GD (1992) Oxidation state-dependent conformational changes in cytochrome c. J Mol Biol 223:959–976
Beasley JR, Pielak GJ (1996) Requirements for perpendicular helix pairing. Proteins 26:95–107
Gochin M, Roder H (1995) Protein structure refinement based on paramagnetic NMR shifts: applications to wild-type and mutant forms of cytochrome c. Protein Sci 4:296–305
Amegadzie BY, Zitomer RS, Hollenberg CP (1990) Characterization of the cytochrome c gene from the starch-fermenting yeast Schwanniomyces occidentalis and its expression in Baker’s yeast. Yeast 6:429–440
Vanfleteren JR, Evers EA, Van de Werken G, Van Beeumen JJ (1990) The primary structure of cytochrome c from the nematode Caenorhabditis elegans. Biochem J 271:613–620
Lyu PC, Liff MI, Marky LA, Kallenbach NR (1990) Side chain contributions to the stability of alpha-helical structure in peptides. Science 250:669–673
Padmanabhan S, Marqusee S, Ridgeway T et al (1990) Relative helix-forming tendencies of nonpolar amino acids. Nature 344:268–270
O’Neil KT, DeGrado WF (1990) A thermodynamic scale for the helix-forming tendencies of the commonly occurring amino acids. Science 250:646–651
Richardson JS, Richardson DC (1988) Amino acid preferences for specific locations at the ends of alpha helices. Science 240:1648–1652
Chothia C, Lesk AM (1985) Helix movements and the reconstruction of the haem pocket during the evolution of the cytochrome c family. J Mol Biol 182:151–158
Burley SK, Petsko GA (1988) Weakly polar interactions in proteins. Adv Protein Chem 39:125–189
Serrano L, Bycroft M, Fersht AR (1991) Aromatic-aromatic interactions and protein stability. Investigation by double-mutant cycles. J Mol Biol 218:465–475
Kleingardner JG, Bren KL (2011) Comparing substrate specificity between cytochrome c maturation and cytochrome c heme lyase systems for cytochrome c biogenesis. Metallomics 3:396–403
Lee I, Salomon AR, Yu K et al (2006) New prospects for an old enzyme: mammalian cytochrome c is tyrosine-phosphorylated in vivo. Biochemistry 45:9121–9128
Garcia-Heredia JM, Diaz-Quintana A, Salzano M et al (2011) Tyrosine phosphorylation turns alkaline transition into a biologically relevant process and makes human cytochrome c behave as an anti-apoptotic switch. J Biol Inorg Chem 16:1155–1168
Creighton TE (1983) An empirical approach to protein conformation stability and flexibility. Biopolymers 22:49–58
Shortle D, Lin B (1985) Genetic analysis of staphylococcal nuclease: identification of three intragenic “global” suppressors of nuclease-minus mutations. Genetics 110:539–555
Das G, Hickey DR, McLendon D et al (1989) Dramatic thermostabilization of yeast iso-1-cytochrome c by an asparagine—isoleucine replacement at position 57. Proc Natl Acad Sci USA 86:496–499
Schweingruber ME, Stewart JW, Sherman F (1979) Primary site and second site revertants of missense mutants of the evolutionarily invariant tryptophan 64 in iso-1-cytochrome c from yeast. J Biol Chem 254:4132–4143
Goto Y, Calciano LJ, Fink AL (1990) Acid-induced folding of proteins. Proc Natl Acad Sci USA 87:573–577
Potekhin S, Pfeil W (1989) Microcalorimetric studies of conformational transitions of ferricytochrome c in acidic solution. Biophys Chem 34:55–62
Stellwagen E, Babul J (1975) Stabilization of the globular structure of ferricytochrome c by chloride in acidic solvents. Biochemistry 14:5135–5140
Nakamura S, Baba T, Kidokoro S (2007) A molten globule-like intermediate state detected in the thermal transition of cytochrome c under low salt concentration. Biophys Chem 127:103–112
Nakamura S, Kidokoro S (2005) Direct observation of the enthalpy change accompanying the native to molten-globule transition of cytochrome c by using isothermal acid-titration calorimetry. Biophys Chem 113:161–168
Sinibaldi F, Howes BD, Smulevich G et al (2003) Anion concentration modulates the conformation and stability of the molten globule of cytochrome c. J Biol Inorg Chem 8:663–670
Sinibaldi F, Piro MC, Howes BD et al (2003) Rupture of the hydrogen bond linking two Omega-loops induces the molten globule state at neutral pH in cytochrome c. Biochemistry 42:7604–7610
Uversky VN, Ptitsyn OB (1994) “Partly folded” state, a new equilibrium state of protein molecules: four-state guanidinium chloride-induced unfolding of beta-lactamase at low temperature. Biochemistry 33:2782–2791
Uversky VN, Ptitsyn OB (1996) Further evidence on the equilibrium “pre-molten globule state”: four-state guanidinium chloride-induced unfolding of carbonic anhydrase B at low temperature. J Mol Biol 255:215–228
Louie GV, Hutcheon WL, Brayer GD (1988) Yeast iso-1-cytochrome c. A 2.8 Å resolution three-dimensional structure determination. J Mol Biol 199:295–314
Tanaka N, Yamane T, Tsukihara T et al (1975) The crystal structure of bonito (katsuo) ferrocytochrome c at 2.3 Å resolution. II. Structure and function. J Biochem 77:147–162
Cookson DJ, Moore GR, Pitt RC et al (1978) Structural homology of cytochromes c. Eur J Biochem 83:261–275
Timkovich R, Dickerson RE (1976) The structure of Paracoccus denitrificans cytochrome c550. J Biol Chem 251:4033–4046
Tsong TY (1974) The Trp-59 fluorescence of ferricytochrome c as a sensitive measure of the over-all protein conformation. J Biol Chem 249:1988–1990
Sherman F, Stewart JW, Jackson M et al (1974) Mutants of yeast defective in iso-1-cytochrome c. Genetics 77:255–284
Lett CM, Guillemette JG (2002) Increasing the redox potential of isoform 1 of yeast cytochrome c through the modification of select haem interactions. Biochem J 362:281–287
Aviram I, Schejter A (1971) Modification of the tryptophanyl residue of horse heart cytochrome c. Biochim Biophys Acta 229:113–118
Caffrey MS, Cusanovich MA (1993) Role of the highly conserved tryptophan of cytochrome c in stability. Arch Biochem Biophys 304:205–208
Black KM, Clark-Lewis I, Wallace CJ (2001) Conserved tryptophan in cytochrome c: importance of the unique side-chain features of the indole moiety. Biochem J 359:715–720
Wallace CJ, Mascagni P, Chait BT et al (1989) Substitutions engineered by chemical synthesis at three conserved sites in mitochondrial cytochrome c. Thermodynamic and functional consequences. J Biol Chem 264:15199–15209
Brayer GD, Murphy MEP (1993) Structural studies of eukaryotic cytochromes c. The Cytochrome c Handbook
Hickey DR, Berghuis AM, Lafond G et al (1991) Enhanced thermodynamic stabilities of yeast iso-1-cytochromes c with amino acid replacements at positions 52 and 102. J Biol Chem 266:11686–11694
Luntz TL, Schejter A, Garber EA, Margoliash E (1989) Structural significance of an internal water molecule studied by site-directed mutagenesis of tyrosine-67 in rat cytochrome c. Proc Natl Acad Sci USA 86:3524–3528
Schejter A, Koshy TI, Luntz TL et al (1994) Effects of mutating Asn-52 to isoleucine on the haem-linked properties of cytochrome c. Biochem J 302(Pt 1):95–101
Schroeder HR, McOdimba FA, Guillemette JG, Kornblatt JA (1997) The polarity of tyrosine 67 in yeast iso-1-cytochrome c monitored by second derivative spectroscopy. Biochem Cell Biol 75:191–197
Berghuis AM, Guillemette JG, McLendon G et al (1994) The role of a conserved internal water molecule and its associated hydrogen bond network in cytochrome c. J Mol Biol 236:786–799
Lett CM, Berghuis AM, Frey HE et al (1996) The role of a conserved water molecule in the redox-dependent thermal stability of iso-1-cytochrome c. J Biol Chem 271:29088–29093
Berghuis AM, Guillemette JG, Smith M, Brayer GD (1994) Mutation of tyrosine-67 to phenylalanine in cytochrome c significantly alters the local heme environment. J Mol Biol 235:1326–1341
Shelnutt JA, Rousseau DL, Dethmers JK, Margoliash E (1981) Protein influences on porphyrin structure in cytochrome c: evidence from Raman difference spectroscopy. Biochemistry 20:6485–6497
Feinberg BA, Liu X, Ryan MD et al (1998) Direct voltammetric observation of redox driven changes in axial coordination and intramolecular rearrangement of the phenylalanine-82-histidine variant of yeast iso-1-cytochrome c. Biochemistry 37:13091–13101
Kassner RJ (1972) Effects of nonpolar environments on the redox potentials of heme complexes. Proc Natl Acad Sci USA 69:2263–2267
Marchon JC, Mashiko T, Reed CA (1982) How does nature control cytochrome redox potentials? In: Ho C et al (eds) Interactions between iron and proteins in oxygen and electron transport, vol 2. Elsevier, pp 67–72
Takano T, Dickerson RE (1981) Conformation change of cytochrome c. II. Ferricytochrome c refinement at 1.8 Å and comparison with the ferrocytochrome structure. J Mol Biol 153:95–115
Mauk AG (1991) Electron transfer in genetically engineered proteins. The cytochrome c paradigm. Struct Bond 75:131–157
Liang N, Pielak GJ, Mauk AG et al (1987) Yeast cytochrome c with phenylalanine or tyrosine at position 87 transfers electrons to (zinc cytochrome c peroxidase)+ at a rate ten thousand times that of the serine-87 or glycine-87 variants. Proc Natl Acad Sci USA 84:1249–1252
Liggins JR, Lo TP, Brayer GD, Nall BT (1999) Thermal stability of hydrophobic heme pocket variants of oxidized cytochrome c. Protein Sci 8:2645–2654
Lo TP, Guillemette JG, Louie GV et al (1995) Structural studies of the roles of residues 82 and 85 at the interactive face of cytochrome c. Biochemistry 34:163–171
Louie GV, Brayer GD (1989) A polypeptide chain-refolding event occurs in the Gly82 variant of yeast iso-1-cytochrome c. J Mol Biol 210:313–322
Louie GV, Pielak GJ, Smith M, Brayer GD (1988) Role of phenylalanine-82 in yeast iso-1-cytochrome c and remote conformational changes induced by a serine residue at this position. Biochemistry 27:7870–7876
Pielak GJ, Oikawa K, Mauk AG et al (1986) Elimination of the negative Soret Cotton effect of eukaryotic cytochromes c by replacement of an invariant phenylalanine residue by site-directed mutagenesis. J Am Chem Soc 108:2724–2727
Rafferty SP, Pearce LL, Barker PD et al (1990) Electrochemical, kinetic, and circular dichroic consequences of mutations at position 82 of yeast iso-1-cytochrome c. Biochemistry 29:9365–9369
Rosell FI, Harris TR, Hildebrand DP et al (2000) Characterization of an alkaline transition intermediate stabilized in the Phe82Trp variant of yeast iso-1-cytochrome c. Biochemistry 39:9047–9054
Torres E, Sandoval JE, Rosell FE et al (1995) Site-directed mutagenesis improves the biocatalytic activity of iso-1-cytochrome c in polycyclic hydrocarbon oxidation. Enzyme Microb Technol 99:779–781
Inglis SC, Guillemette JG, Johnson JA, Smith M (1991) Analysis of the invariant Phe82 residue of yeast iso-1-cytochrome c by site-directed mutagenesis using a phagemid yeast shuttle vector. Protein Eng 4:569–574
Pielak GJ, Mauk AG, Smith M (1985) Site-directed mutagenesis of cytochrome c shows that an invariant Phe is not essential for function. Nature 313:152–154
Liang N, Mauk AG, Pielak GJ et al (1988) Regulation of interprotein electron transfer by residue 82 of yeast cytochrome c. Science 240:311–313
Poulos TL, Kraut J (1980) A hypothetical model of the cytochrome c peroxidase. cytochrome c electron transfer complex. J Biol Chem 255:10322–10330
Wendoloski JJ, Matthew JB, Weber PC, Salemme FR (1987) Molecular dynamics of a cytochrome c-cytochrome b5 electron transfer complex. Science 238:794–797
Pearce LL, Gartner AL, Smith M, Mauk AG (1989) Mutation-induced perturbation of the cytochrome c alkaline transition. Biochemistry 28:3152–3156
Ochi H, Hata Y, Tanaka N et al (1983) Structure of rice ferricytochrome c at 2.0 Å resolution. J Mol Biol 166:407–418
Pielak GJ, Concar DW, Moore GR, Williams RJ (1987) The structure of cytochrome c and its relation to recent studies of long-range electron transfer. Protein Eng 1:83–88
Greene RM, Betz SF, Hilgen-Willis S et al (1993) Changes in global stability and local structure of cytochrome c upon substituting phenylalanine-82 with tyrosine. J Inorg Biochem 51:663–676
Lum VR, Brayer GD, Louie GV et al (1987) Protein Struct. Fold Design 26:143–150
Dickerson RE (1980) Cytochrome c and the evolution of energy metabolism. Sci Am 242:137–153
Summers MR, McPhie P (1972) The mechanism of unfolding of globular proteins. Biochem Biophys Res Commun 47:831–837
Tsong TY (1973) Detection of three kinetic phases in the thermal unfolding of ferricytochrome c. Biochemistry N10I:2209–2214
Brandts JF, Halvorson HR, Brennan M (1975) Consideration of the Possibility that the slow step in protein denaturation reactions is due to cis-trans isomerism of proline residues. Biochemistry 14:4953–4963
Ramdas L, Nall BT (1986) Folding/unfolding kinetics of mutant forms of iso-1-cytochrome c with replacement of proline-71. Biochemistry 25:6959–6964
White TB, Berget PB, Nall BT (1987) Changes in conformation and slow refolding kinetics in mutant iso-2-cytochrome c with replacement of a conserved proline residue. Biochemistry 26:4358–4366
Koshy TI, Luntz TL, Schejter A, Margoliash E (1990) Changing the invariant proline-30 of rat and Drosophila melanogaster cytochromes c to alanine or valine destabilizes the heme crevice more than the overall conformation. Proc Natl Acad Sci USA 87:8697–8701
George P, Lyster RL (1958) Crevice structures in hemoprotein reactions. Proc Natl Acad Sci USA 44:1013–1029
Baistrocchi P, Banci L, Bertini I et al (1996) Three-dimensional solution structure of Saccharomyces cerevisiae reduced iso-1-cytochrome c. Biochemistry 35:13788–13796
Kellis JT Jr, Nyberg K, Sali D, Fersht AR (1988) Contribution of hydrophobic interactions to protein stability. Nature 333:784–786
Poerio E, Parr GR, Taniuchi H (1986) A study of roles of evolutionarily invariant proline 30 and glycine 34 of cytochrome c. J Biol Chem 261:10976–10989
Wood LC, Muthukrishnan K, White TB et al (1988) Construction and characterization of mutant iso-2-cytochromes c with replacement of conserved prolines. Biochemistry 27:8554–8561
Gooley PR, MacKenzie NE (1990) Pro—Ala-35 Rhodobacter capsulatus cytochrome c2 shows dynamic not structural differences. A 1H and 15N NMR study. FEBS Lett 260:225–228
Lan W, Wang Z, Yang Z et al (2011) Conformational toggling of yeast iso-1-cytochrome C in the oxidized and reduced states. PLoS ONE 6:e27219
Ernst JF, Hampsey DM, Stewart JW et al (1985) Substitutions of proline 76 in yeast iso-1-cytochrome c. Analysis of residues compatible and incompatible with folding requirements. J Biol Chem 260:13225–13236
Wallace CJA, Lewis IC (1997) A rationale for the absolute conservation of Asn 70 and pro in mitochondrial cytochromes c suggested by protein engineering. Biochemistry 39:395–399
Ernst JF, Stewart JW, Sherman F (1981) The cyc1-11 mutation in yeast reverts by recombination with a nonallelic gene: composite genes determining the iso-cytochromes c. Proc Natl Acad Sci USA 78:6334–6338
Ramdas L, Sherman F, Nall BT (1986) Guanidine hydrochloride induced equilibrium unfolding of mutant forms of iso-1-cytochrome c with replacement of proline-71. Biochemistry 25:6952–6958
Wood LC, White TB, Ramdas L, Nall BT (1988) Replacement of a conserved proline eliminates the absorbance-detected slow folding phase of iso-2-cytochrome c. Biochemistry 27:8562–8568
Murphy MEP (1993) Ph.D. Dissertation, University of British Columbia
Wuttke DS, Gray HB (1993) Protein engineering as a tool for understanding electron transfer. Curr Opin Struct Biol 3:555–563
Mulligan-Pullyblank P, Spitzer JS, Gilden BM, Fetrow JS (1996) Loop replacement and random mutagenesis of omega-loop D, residues 70–84, in iso-1-cytochrome c. J Biol Chem 271:8633–8645
Black KM, Wallace CJ (2007) Probing the role of the conserved beta-II turn Pro-76/Gly-77 of mitochondrial cytochrome c. Biochem Cell Biol 85:366–374
Fetrow JS, Spitzer JS, Gilden BM et al (1998) Structure, function, and temperature sensitivity of directed, random mutants at proline 76 and glycine 77 in omega-loop D of yeast iso-1-cytochrome c. Biochemistry 37:2477–2487
Ahmed AJ, Smith HT, Smith MB, Millett FS (1978) Effect of specific lysine modification on the reduction of cytochrome c by succinate-cytochrome c reductase. Biochemistry 17:2479–2483
Ferguson-Miller S, Brautigan DL, Margoliash E (1978) Definition of cytochrome c binding domains by chemical modification. III. Kinetics of reaction of carboxydinitrophenyl cytochromes c with cytochrome c oxidase. J Biol Chem 253:149–159
Koppenol WH, Margoliash E (1982) The asymmetric distribution of charges on the surface of horse cytochrome c. Functional implications. J Biol Chem 257:4426–4437
Rieder R, Bosshard HR (1978) The cytochrome c oxidase binding site on cytochrome c. Differential chemical modification of lysine residues in free and oxidase-bound cytochrome c. J Biol Chem 253:6045–6053
DeLange RJ, Glazer AN, Smith EL (1970) Identification and location of episilon-N-trimethyllysine in yeast cytochromes c. J Biol Chem 245:3325–3327
Paik WK, Cho YB, Frost B, Kim S (1989) Cytochrome c methylation. Biochem Cell Biol 67:602–611
Takakura H, Yamamoto T, Sherman F (1997) Sequence requirement for trimethylation of yeast cytochrome c. Biochemistry 36:2642–2648
Holzschu D, Principio L, Conklin KT et al (1987) Replacement of the invariant lysine 77 by arginine in yeast iso-1-cytochrome c results in enhanced and normal activities in vitro and in vivo. J Biol Chem 262:7125–7131
Sharonov GV, Feofanov AV, Bocharova OV et al (2005) Comparative analysis of proapoptotic activity of cytochrome c mutants in living cells. Apoptosis 10:797–808
Kluck RM, Ellerby LM, Ellerby HM et al (2000) Determinants of cytochrome c pro-apoptotic activity. The role of lysine 72 trimethylation. J Biol Chem 275:16127–16133
Polastro E, Looze Y, Leonis J (1976) Evidence that trimethylation of iso-1-cytochrome c from Saccharomyces cerevisiae does not alter its functional properties [proceedings]. Arch Int Physiol Biochim 84:1099–1100
Polastro E, Looze Y, Leonis J (1976) Study of the biological significance of cytochrome methylation. I. Thermal, acid and guanidinium hydrochloride denaturations of baker’s yeast ferricytochromes c. Biochim Biophys Acta 446:310–320
Polastro E, Looze Y, Leonis J (1976) Study of the biological significance of the methylation of cytochromes c. I. Thermal, acid and guanidinium hydrochloride denaturations of baker’s yeast ferricytochromes c. Arch Int Physiol Biochim 84:407–409
Polastro ET, Deconinck MM, Devogel MR et al (1978) Evidence that trimethylation of iso-I-cytochrome c from Saccharomyces cerevisiae affects interaction with the mitochondrion. FEBS Lett 86:17–20
Guiard B, Lederer F (1979) The “cytochrome b5 fold”: structure of a novel protein superfamily. J Mol Biol 135:639–650
Paik WK, Polastro E, Kim S (1980) Cytochrome c methylation: enzymology and biologic significance. Curr Top Cell Regul 16:87–111
Frost B, Park KS, Kim S, Paik WK (1989) Effect of enzymatic methylation of apocytochrome c on holocytochrome c formation and proteolysis. Int J Biochem 21:1407–1414
Li P, Nijhawan D, Budihardjo I et al (1997) Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell 91:479–489
Liu X, Kim CN, Yang J et al (1996) Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell 86:147–157
Rodrigues J, Lazebnic Y (1999) Caspase-9 and APAF-1 form an active holoenzyme. Genes Dev 13:3179–3184
Chertkova RV, Sharonov GV, Feofanov AV et al (2008) Proapoptotic activity of cytochrome c in living cells: effect of K72 substitutions and species differences. Mol Cell Biochem 314:85–93
Yu T, Wang X, Purring-Koch C et al (2001) A mutational epitope for cytochrome C binding to the apoptosis protease activation factor-1. J Biol Chem 276:13034–13038
Cortese JD, Voglino AL, Hackenbrock CR (1998) Multiple conformations of physiological membrane-bound cytochrome c. Biochemistry 37:6402–6409
Heimburg T, Marsh D (1995) Protein surface-distribution and protein-protein interactions in the binding of peripheral proteins to charged lipid membranes. Biophys J 68:536–546
Nicholls P (1974) Cytochrome c binding to enzymes and membranes. Biochim Biophys Acta 346:261–310
Pinheiro TJ, Elove GA, Watts A, Roder H (1997) Structural and kinetic description of cytochrome c unfolding induced by the interaction with lipid vesicles. Biochemistry 36:13122–13132
Pinheiro TJ, Watts A (1994) Lipid specificity in the interaction of cytochrome c with anionic phospholipid bilayers revealed by solid-state 31P NMR. Biochemistry 33:2451–2458
Soussi B, Bylund-Fellenius AC, Schersten T, Angstrom J (1990) 1H-n.m.r. evaluation of the ferricytochrome c-cardiolipin interaction. Effect of superoxide radicals. Biochem J 265:227–232
Zhang F, Rowe ES (1994) Calorimetric studies of the interactions of cytochrome c with dioleoylphosphatidylglycerol extruded vesicles: ionic strength effects. Biochim Biophys Acta 1193:219–225
Dickerson RE, Timkovich R (1975) The Enzymes (Boyer, P. D., ed), vol 11. Academic Press, Orlando, pp 397–547
Yu H, Lee I, Salomon AR et al (2008) Mammalian liver cytochrome c is tyrosine-48 phosphorylated in vivo, inhibiting mitochondrial respiration. Biochim Biophys Acta 1777:1066–1071
Ischiropoulos H (2003) Biological selectivity and functional aspects of protein tyrosine nitration. Biochem Biophys Res Commun 305:776–783
Ischiropoulos H (2009) Protein tyrosine nitration–an update. Arch Biochem Biophys 484:117–121
Souza JM, Castro L, Cassina AM et al (2008) Nitrocytochrome c: synthesis, purification, and functional studies. Methods Enzymol 441:197–215
Batthyany C, Souza JM, Duran R et al (2005) Time course and site(s) of cytochrome c tyrosine nitration by peroxynitrite. Biochemistry 44:8038–8046
Cassina AM, Hodara R, Souza JM et al (2000) Cytochrome c nitration by peroxynitrite. J Biol Chem 275:21409–21415
Jang B, Han S (2006) Biochemical properties of cytochrome c nitrated by peroxynitrite. Biochimie 88:53–58
MacMillan-Crow LA, Cruthirds DL, Ahki KM et al (2001) Mitochondrial tyrosine nitration precedes chronic allograft nephropathy. Free Radic Biol Med 31:1603–1608
Nakagawa H, Komai N, Takusagawa M et al (2007) Nitration of specific tyrosine residues of cytochrome C is associated with caspase-cascade inactivation. Biol Pharm Bull 30:15–20
Rodriguez-Roldan V, Garcia-Heredia JM, Navarro JA et al (2008) Effect of nitration on the physicochemical and kinetic features of wild-type and monotyrosine mutants of human respiratory cytochrome c. Biochemistry 47:12371–12379
Souza JM, Peluffo G, Radi R (2008) Protein tyrosine nitration–functional alteration or just a biomarker? Free Radic Biol Med 45:357–366
Ueta E, Kamatani T, Yamamoto T, Osaki T (2003) Tyrosine-nitration of caspase 3 and cytochrome c does not suppress apoptosis induction in squamous cell carcinoma cells. Int J Cancer 103:717–722
Diaz-Moreno I, Garcia-Heredia JM, Diaz-Quintana A et al (2011) Nitration of tyrosines 46 and 48 induces the specific degradation of cytochrome c upon change of the heme iron state to high-spin. Biochim Biophys Acta 1807:1616–1623
Castro L, Eiserich JP, Sweeney S et al (2004) Cytochrome c: a catalyst and target of nitrite-hydrogen peroxide-dependent protein nitration. Arch Biochem Biophys 421:99–107
Chen YR, Chen CL, Chen W et al (2004) Formation of protein tyrosine ortho-semiquinone radical and nitrotyrosine from cytochrome c-derived tyrosyl radical. J Biol Chem 279:18054–18062
Basova LV, Kurnikov IV, Wang L et al (2007) Cardiolipin switch in mitochondria: shutting off the reduction of cytochrome c and turning on the peroxidase activity. Biochemistry 46:3423–3434
Nakagawa D, Ohshima Y, Takusagawa M et al (2001) Functional modification of cytochrome c by peroxynitrite in an electron transfer reaction. Chem Pharm Bull (Tokyo) 49:1547–1554
Florence TM (1985) The degradation of cytochrome c by hydrogen peroxide. J Inorg Biochem 23:131–141
Radi R, Thomson L, Rubbo H, Prodanov E (1991) Cytochrome c-catalyzed oxidation of organic molecules by hydrogen peroxide. Arch Biochem Biophys 288:112–117
Chen YR, Deterding LJ, Sturgeon BE et al (2002) Protein oxidation of cytochrome C by reactive halogen species enhances its peroxidase activity. J Biol Chem 277:29781–29791
Kagan VE, Bayir HA, Belikova NA et al (2009) Cytochrome c/cardiolipin relations in mitochondria: a kiss of death. Free Radic Biol Med 46:1439–1453
Oursler MJ, Bradley EW, Elfering SL, Giulivi C (2005) Native, not nitrated, cytochrome c and mitochondria-derived hydrogen peroxide drive osteoclast apoptosis. Am J Physiol Cell Physiol 288:C156–C168
Abriata LA, Cassina A, Tortora V et al (2009) Nitration of solvent-exposed tyrosine 74 on cytochrome c triggers heme iron-methionine 80 bond disruption. Nuclear magnetic resonance and optical spectroscopy studies. J Biol Chem 284:17–26
Giulivi C, Poderoso JJ, Boveris A (1998) Production of nitric oxide by mitochondria. J Biol Chem 273:11038–11043
Radi R, Cassina A, Hodara R et al (2002) Peroxynitrite reactions and formation in mitochondria. Free Radic Biol Med 33:1451–1464
Kim SC, Sprung R, Chen Y et al (2006) Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell 23:607–618
Azzi A, Montecucco C, Richter C (1975) The use of acetylated ferricytochrome c for the detection of superoxide radicals produced in biological membranes. Biochem Biophys Res Commun 65:597–603
Arnesen T (2011) Towards a functional understanding of protein N-terminal acetylation. PLoS Biol 9:e1001074
Polevoda B, Norbeck J, Takakura H et al (1999) Identification and specificities of N-terminal acetyltransferases from Saccharomyces cerevisiae. EMBO J 18:6155–6168
Polevoda B, Sherman F (2000) Nalpha -terminal acetylation of eukaryotic proteins. J Biol Chem 275:36479–36482
Hershko A, Heller H, Eytan E et al (1984) Role of the alpha-amino group of protein in ubiquitin-mediated protein breakdown. Proc Natl Acad Sci USA 81:7021–7025
Matsuura S, Arpin M, Hannum C et al (1981) In vitro synthesis and posttranslational uptake of cytochrome c into isolated mitochondria: role of a specific addressing signal in the apocytochrome. Proc Natl Acad Sci USA 78:4368–4372
Laz TM, Pietras DF, Sherman F (1984) Differential regulation of the duplicated isocytochrome c genes in yeast. Proc Natl Acad Sci USA 81:4475–4479
Prezant T, Pfeifer K, Guarente L (1987) Organization of the regulatory region of the yeast CYC7 gene: multiple factors are involved in regulation. Mol Cell Biol 7:3252–3259
Zhang Z, Gerstein M (2003) The human genome has 49 cytochrome c pseudogenes, including a relic of a primordial gene that still functions in mouse. Gene 312:61–72
Limbach KJ, Wu R (1985) Characterization of two Drosophila melanogaster cytochrome c genes and their transcripts. Nucleic Acids Res 13:631–644
Swanson MS, Zieminn SM, Miller DD et al (1985) Developmental expression of nuclear genes that encode mitochondrial proteins: insect cytochromes c. Proc Natl Acad Sci USA 82:1964–1968
Kim IC, Nolla H (1986) Antigenic analysis of testicular cytochromes c using monoclonal antibodies. Biochem Cell Biol 64:1211–1217
Kim IC, Sabourin CL (1986) Antigenic and size differences between somatic and testicular cytochromes c. Biochem Biophys Res Commun 141:131–136
Scarpulla RC, Agne KM, Wu R (1981) Isolation and structure of a rat cytochrome c gene. J Biol Chem 256:6480–6486
Scarpulla RC (1984) Processed pseudogenes for rat cytochrome c are preferentially derived from one of three alternate mRNAs. Mol Cell Biol 4:2279–2288
Huttemann M, Jaradat S, Grossman LI (2003) Cytochrome c oxidase of mammals contains a testes-specific isoform of subunit VIb–the counterpart to testes-specific cytochrome c? Mol Reprod Dev 66:8–16
Narisawa S, Hecht NB, Goldberg E et al (2002) Testis-specific cytochrome c-null mice produce functional sperm but undergo early testicular atrophy. Mol Cell Biol 22:5554–5562
Virbasius JV, Scarpulla RC (1988) Structure and expression of rodent genes encoding the testis-specific cytochrome c. Differences in gene structure and evolution between somatic and testicular variants. J Biol Chem 263:6791–6796
Hennig B (1975) Change of cytochrome c structure during development of the mouse. Eur J Biochem 55:167–183
Davies AM, Guillemette JG, Smith M et al (1993) Redesign of the interior hydrophilic region of mitochondrial cytochrome c by site-directed mutagenesis. Biochemistry 32:5431–5435
Cadenas E, Davies KJ (2000) Mitochondrial free radical generation, oxidative stress, and aging. Free Radic Biol Med 29:222–230
Han D, Williams E, Cadenas E (2001) Mitochondrial respiratory chain-dependent generation of superoxide anion and its release into the intermembrane space. Biochem J 353:411–416
Han D, Canali R, Rettori D, Kaplowitz N (2003) Effect of glutathione depletion on sites and topology of superoxide and hydrogen peroxide production in mitochondria. Mol Pharmacol 64:1136–1144
Kadenbach B, Arnold S, Lee I, Huttemann M (2004) The possible role of cytochrome c oxidase in stress-induced apoptosis and degenerative diseases. Biochim Biophys Acta 1655:400–408
Turrens JF, Alexandre A, Lehninger AL (1985) Ubisemiquinone is the electron donor for superoxide formation by complex III of heart mitochondria. Arch Biochem Biophys 237:408–414
Aoki H, Kang PM, Hampe J et al (2002) Direct activation of mitochondrial apoptosis machinery by c-Jun N-terminal kinase in adult cardiac myocytes. J Biol Chem 277:10244–10250
Acknowledgments
We sincerely thank our reviewers for their critical reading and helpful suggestions. FA thanks the Department of Science and Technology (India) for financial support. SZ and MIH are thankful to the Indian Council of Medical Research for fellowship and grants, respectively. We apologize to those whose work was not cited or insufficiently cited because of space limitations.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zaidi, S., Hassan, M.I., Islam, A. et al. The role of key residues in structure, function, and stability of cytochrome-c . Cell. Mol. Life Sci. 71, 229–255 (2014). https://doi.org/10.1007/s00018-013-1341-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-013-1341-1