Abstract
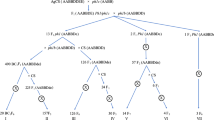
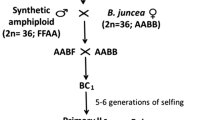
A set of 110 diploid putative introgression lines (ILs) containing chromatin introgressed from the undomesticated species Hordeum bulbosum L. (bulbous barley grass) into cultivated barley (Hordeum vulgare L.) has been identified using a high-copy number retrotransposon-like PCR marker, pSc119.1, derived from rye (Secale cereale L.). To evaluate these lines, 92 EST-derived markers were developed by marker sequencing across four barley cultivars and four H. bulbosum genotypes. Single nucleotide polymorphisms and insertions/deletions conserved between the two species were then used to develop a set of fully informative cleaved amplified polymorphic sequence markers or size polymorphic insertion/deletion markers. Introgressed chromatin from H. bulbosum was confirmed and genetically located in 88 of these lines using 46 of the EST-derived PCR markers. A total of 96 individual introgressions were detected with most of them (94.8%) extending to the most distal marker for each respective chromosome arm. Introgressions were detected on all chromosome arms except chromosome 3HL. Interstitial or sub-distal introgressions also occurred, with two located on chromosome 2HL and one each on 3HS, 5HL and 6HS. Twenty-two putative ILs that were positive for H. bulbosum chromatin using pSc119.1 have not had introgressions detected with these single-locus markers. When all introgressions are combined, more than 36% of the barley genetic map has now been covered with introgressed chromatin from H. bulbosum. These ILs represent a significant germplasm resource for barley improvement that can be mined for diverse traits of interest to barley breeders and researchers.



Similar content being viewed by others
References
Badr A, Muller K, Schafer-Pregl R, El Rabey H, Effgen S, Ibrahim H, Pozzi C, Rohde W, Salamini F (2000) On the origin and domestication history of barley (Hordeum vulgare). Mol Biol Evol 17:499–510
Chiapparino E, Lee D, Donini P (2004) Genotyping single nucleotide polymorphisms in barley by tetra-primer ARMS-PCR. Genome 47:414–420
Feuillet C, Langridge P, Waugh R (2008) Cereal breeding takes a walk on the wild side. Trends Genet 24:24–32
GenStat Committee (2006) The guide to GenStat Release 9—parts 1–3. In: Payne R (ed) VSN International, Oxford
Ilic K, Berleth T, Provart N (2004) BlastDigester—a web-based program for efficient CAPS marker design. Trends Genet 20:280–283
Jaffé B, Caligari P, Snape J (2000) A skeletal map of Hordeum bulbosum L. and comparative mapping with barley. Euphytica 115:115–120
Johnston P (2007) Molecular characterisation of chromatin introgressed from Hordeum bulbosum L. into Hordeum vulgare L. PhD Thesis, Department of Biochemistry. University of Otago, Dunedin, New Zealand, p 340
Johnston P, Pickering R (2002) PCR detection of Hordeum bulbosum introgressions in an H. vulgare background using a retrotransposon-like sequence. Theor Appl Genet 104:720–726
Kleinhofs A, Killian A, Saghai-Maroof M, Biyashev R, Hayes P, Chen F, Lapitan N, Fenwick A, Blake T, Kanazin V, Ananiev E, Dahleen L, Kudrna D, Bollinger J, Knapp S, Liu B, Sorrells M, Heun M, Franckowiak J, Hoffman D, Skadsen R, Steffenson B (1993) A molecular, isozyme and morphological map of the barley (Hordeum vulgare) genome. Theor Appl Genet 86:705–712
Korzun V, Malyshev S, Pickering R, Borner A (1999) RFLP mapping of a gene for hairy leaf sheath using a recombinant line from Hordeum vulgare L. × Hordeum bulbosum L. cross. Genome 42:960–963
Kota R, Varshney R, Prasad M, Zhang H, Stein N, Graner A (2007) EST-derived single nucleotide polymorphism markers for assembling genetic and physical maps of the barley genome. Funct Integr Genomics 8:223–233
Künzel G, Korzun L, Meister A (2000) Cytologically integrated physical restriction fragment length polymorphism maps for the barley genome based on translocation breakpoints. Genetics 154:397–412
Manly K, Cudmore R, Meer J (2001) Map Manager QTX, cross-platform software for genetic mapping. Mamm Genome 12:930–932
McCullagh P, Nelder J (1989) Generalized linear models, 2nd edn. Chapman and Hall, London
Murray M, Thompson W (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4326
Palumbi S (1995) Nucleic acids II: the polymerase chain reaction. In: Hillis D, Moritz C (eds) Molecular systematics, 2nd edn. MA Sinauer Associates Inc., Sunderland, pp 205–247
Parmley JL, Urrutia AO, Potrzebowski L, Kaessmann H, Hurst LD (2007) Splicing and the evolution of proteins in mammals. PLoS Biol 5:e14
Pickering R (1988) The production of fertile triploid hybrids between Hordeum vulgare L. (2n = 2x = 14) and H. bulbosum L. (2n = 4x = 28). Barley Genet Newsl 18:25–29
Pickering R (2000) Do the wild relatives of cultivated barley have a place in barley improvement? In: Logue S (ed) Barley genetics. In: VIII Proceedings of the 8th international barley genetics symposium, Adelaide, Australia, pp 223–230
Pickering R, Hill A, Michel M, Timmerman-Vaughan G (1995) The transfer of a powdery mildew resistance gene from Hordeum bulbosum L. to barley (H. vulgare L.) chromosome 2 (2I). Theor Appl Genet 91:1288–1292
Pickering R, Steffenson B, Hill A, Borovkova I (1998) Association of leaf rust and powdery mildew resistance in a recombinant derived from a Hordeum vulgare × Hordeum bulbosum hybrid. Plant Breed 117:83–84
Pickering R, Malyshev S, Kunzel G, Johnston P, Korzun V, Menke M, Schubert I (2000) Locating introgressions of Hordeum bulbosum chromatin within the H. vulgare genome. Theor Appl Genet 100:27–31
Pickering R, Klatte S, Butler R (2006) Identification of all chromosome arms and their involvement in meiotic homoeologous associations at metaphase I in 2 Hordeum vulgare L. × Hordeum bulbosum L. hybrids. Genome 49:73–78
Qi LL, Echalier B, Chao S, Lazo GR, Butler GE, Anderson OD, Akhunov ED, Dvorak J, Linkiewicz AM, Ratnasiri A, Dubcovsky J, Bermudez-Kandianis CE, Greene RA, Kantety R, La Rota CM, Munkvold JD, Sorrells SF, Sorrells ME, Dilbirligi M, Sidhu D, Erayman M, Randhawa HS, Sandhu D, Bondareva SN, Gill KS, Mahmoud AA, Ma XF, Miftahudin, Gustafson JP, Conley EJ, Nduati V, Gonzalez-Hernandez JL, Anderson JA, Peng JH, Lapitan NLV, Hossain KG, Kalavacharla V, Kianian SF, Pathan MS, Zhang DS, Nguyen HT, Choi DW, Fenton RD, Close TJ, McGuire PE, Qualset CO, Gill BS (2004) A chromosome bin map of 16,000 expressed sequence tag loci and distribution of genes among the three genomes of polyploid wheat. Genetics 168:701–712
Ramsay L, Macaulay M, degli Ivanissevich S, MacLean K, Cardle L, Fuller J, Edwards KJ, Tuvesson S, Morgante M, Massari A, Maestri E, Marmiroli N, Sjakste T, Ganal M, Powell W, Waugh R (2000) A simple sequence repeat-based linkage map of barley. Genetics 156:1997–2005
Rostoks N, Mudie S, Cardle L, Russell J, Ramsay L, Booth A, Svensson JT, Wanamaker SI, Walia H, Rodriguez EM, Hedley P, Liu H, Morris J, Close TJ, Marshall DF, Waugh R (2005) Genome-wide SNP discovery and linkage analysis in barley based on genes responsive to abiotic stress. Mol Genet Genomics 274:515–527
Ruge B, Linz A, Pickering R, Proeseler G, Greif P, Wehling P (2003) Mapping of Rym14 Hb, a gene introgressed from Hordeum bulbosum and conferring resistance to BaMMV and BaYMV in barley. Theor Appl Genet 107:965–971
Salvo-Garrido H, Laurie D, Jaffe B, Snape J (2001) An RFLP map of diploid Hordeum bulbosum L. and comparison with maps of barley (H. vulgare L.) and wheat (Triticum aestivum L.). Theor Appl Genet 103:869–880
Sato K, Nankaku N, Motoi Y, Takeda K (2004) A large scale mapping of ESTs on barley genome. In: Spunar J, Janikova J (eds) 9th International barley genetics symposium, Brno, Czech Republic
Szigat G, Pohler W (1982) Hordeum bulbosum × H. vulgare hybrids and their backcrosses with cultivated barley. Cereal Res Commun 10:73–78
Thiel T, Michalek W, Varshney R, Graner A (2003) Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor Appl Genet 106:411–422
Toubia-Rahme H, Johnston P, Pickering R, Steffenson B (2003) Inheritance and chromosomal location of Septoria passerinii resistance introgressed from Hordeum bulbosum into Hordeum vulgare. Plant Breed 122:405–409
Varshney RK, Marcel TC, Ramsay L, Russell J, Röder MS, Stein N, Waugh R, Langridge P, Niks RE, Graner A (2007) A high density barley microsatellite consensus map with 775 SSR loci. Theor Appl Genet 114:1091–1103
von Bothmer R, Jacobsen N, Baden C, Jorgensen R, Linde-Laursen I (1995) An ecogeographical study of the genus Hordeum, 2nd edn. International Plant Genetic Resources Institute (IPGRI), Rome
Walther U, Rapke H, Proeseler G, Szigat G (2000) Hordeum bulbosum—a new source of disease resistance—transfer of resistance to leaf rust and mosaic viruses from H. bulbosum into winter barley. Plant Breed 119:215–218
Wanamaker S, Close T, Roose M, Lyon M (2006) HarvEST: Barley EST database version 1.44; http://harvest.ucr.edu/. Cited 10 November 2006
Xu J, Kasha KJ (1992) Transfer of a dominant gene for powdery mildew resistance and DNA from Hordeum bulbosum into cultivated barley (H. vulgare). Theor Appl Genet 84:771–777
Zhu YL, Song QJ, Hyten DL, Van Tassell CP, Matukumalli LK, Grimm DR, Hyatt SM, Fickus EW, Young ND, Cregan PB (2003) Single-nucleotide polymorphisms in soybean. Genetics 163:1123–1134
Acknowledgments
The authors would like to thank Professor Kazuhiro Sato for providing primer sequences of mapped EST markers, Mrs. Ruth Butler for statistical analysis of SNP frequencies and Dr. Samantha Baldwin for helping construct the figures for this manuscript. Comments from journal reviewers made a significant improvement to this manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Graner.
Electronic supplementary material
Below is the link to the electronic supplementary material.
122_2009_992_MOESM1_ESM.xls
Supplementary Table S1 A full list of the plant materials used in this study including information on pedigree, known traits derived from H. bulbosum and the chromosomal location of any detected introgressions (XLS 40 kb)
122_2009_992_MOESM2_ESM.xls
Supplementary Table S2 PCR primers and conditions for all the markers used in this study and including the diagnostic tools (restriction enzymes or size polymorphisms) used to discriminate between barley and H. bulbosum alleles. The restriction enzymes used for genetic mapping in the Steptoe x Morex population (Kleinhofs et al. 1993) are also included (XLS 59 kb)
Rights and permissions
About this article
Cite this article
Johnston, P.A., Timmerman-Vaughan, G.M., Farnden, K.J.F. et al. Marker development and characterisation of Hordeum bulbosum introgression lines: a resource for barley improvement. Theor Appl Genet 118, 1429–1437 (2009). https://doi.org/10.1007/s00122-009-0992-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-009-0992-7