Abstract
The proteome of extremely thermophilic microorganisms affords a glimpse into the dynamics of microbial ecology of high temperature environments. The secretome, or extracellular proteome of these microorganisms, no doubt harbors technologically important enzymes and other thermostable biomolecules that, to date, have been characterized only to a limited extent. In the first of a two-part study on selected thermophiles, defining the secretome requires a sample preparation method that has no negative impact on all downstream experiments. Following efficient secretome purification, GeLC-MS2 analysis and prediction servers suggested probable protein secretion to complement experimental data. In an effort to define the extracellular proteome of the extreme thermophilic bacterium Caldicellulosiruptor saccharolyticus, several techniques were considered regarding sample processing to achieve the most in-depth analysis of secreted proteins. Order of operation experiments, all including the C18 bead technique, demonstrated that two levels of sample purification were necessary to effectively desalt the sample and provide sufficient protein identifications. Five sample preparation combinations yielded 71 proteins and the majority described, as enzymatic and putative uncharacterized proteins, anticipate consolidated bioprocessing applications. Nineteen proteins were predicted by Phobius, SignalP, SecretomeP, or TatP for extracellular secretion, and 11 contained transmembrane domain stretches suggested by Phobius and transmembrane hidden Markov model. The sample preparation technique demonstrating the most effective outcome for C. saccharolyticus secreted proteins in this study, involved acetone precipitation followed by the C18 bead method in which 2.4% (63 proteins) of the predicted proteome was identified, including proteins suggested to have secretion and transmembrane moieties.

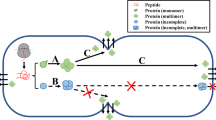
Experimental workflow for the evaluation of sample cleanup techniques for the secretome of the thermophilic bacterium Caldicellulosiruptor saccharolyticus with possibility to traverse other similarly grown bacterium. Several sample purification methods were assessed individually as well as in combination with a C18 bead method in an effort to afford the greatest number of confidently identified proteins. Analysis of the identified proteins by prediction servers complemented the experimental secretome investigation.




Similar content being viewed by others
References
Antranikian G, Egorova K (2007) Extremophiles, a unique resource of biocatalysts for industrial biotechnology. In: Gerday C, Glansdorff N (eds) Physiology and biochemistry of extremophiles. ASM Press, Washington, pp 361–406
VanFossen AL, Lewis DL, Nichols JD, Kelly RM (2008) Polysaccharide degradation and synthesis by extremely nermophilic anaerobes. Ann NY Acad Sci 1125:322–337
Vafiadi C, Topakas E, Biely P, Christakopoulos P (2009) Purification, characterization and mass spectrometric sequencing of a thermophilic glucuronoyl esterase from Sporotrichum thermophile. FEMS Microbiol Lett 296:178–184
Barabote RD, Xie G, Leu DH, Normand P, Necsulea A, Daubin V, Medigue C, Adney WS, Xu XC, Lapidus A, Parales RE, Detter C, Pujic P, Bruce D, Lavire C, Challacombe JF, Brettin TS, Berry AM (2009) Complete genome of the cellulolytic thermophile Acidothermus cellulolyticus 11B provides insights into its ecophysiological and evolutionary adaptations. Genome Res 19:1033–1043
Asada Y, Endo S, Inoue Y, Mamiya H, Hara A, Kunishima N, Matsunaga T (2009) Biochemical and structural characterization of a short-chain dehydrogenase/reductase of Thermus thermophilus HB8 A hyperthermostable aldose-1-dehydrogenase with broad substrate specificity. Chem Biol Interact 178:117–126
Muhammad SA, Ahmad S, Hameed A (2009) Antibiotic production by Thermophilic bacillus specie Sat-4. Pak J Pharm Sci 22:339–345
Esikova TZ, Temirov YV, Sokolov SL, Alakhov YB (2002) Secondary antimicrobial metabolites produced by thermophilic Bacillus spp. strains VK2 and VK21. Appl Biochem Microbiol 38:226–231
Sato S, Hutchison CA, Harris JI (1977) Thermostable sequence-specific endonuclease from Thermus aquaticus. PNAS 74:542–546
Tjalsma H, Bolhuis A, Jongbloed JDH, Bron S, van Dijl JM (2000) Signal peptide-dependent protein transport in Bacillus subtilis: a genome-based survey of the secretome. Microbiol Mol Biol Rev 64:515–547
Zhou M, Boekhorst J, Francke C, Siezen RJ (2008) LocateP: genome-scale subcellular-location predictor for bacterial proteins. BMC Bioinform 9:173
Hathout Y (2007) Approaches to the study of the cell secretome. Expert Rev Proteomics 4:239–248
Rainey FA, Donnison AM, Janssen PH, Saul D, Rodrigo A, Bergquist PL, Daniel RM, Stackebrandt E, Morgan HW (1994) Description of Caldicellulosiruptor saccharolyticus gen. nov., sp. nov: an obligately anaerobic, extremely thermophilic, cellulolytic bacterium. FEMS Microbiol Lett 120:263–266
Blumer-Schuette SE, Kataeva I, Westpheling J, Adams MWW, Kelly RM (2008) Extremely thermophilic microorganisms for biomass conversion: status and prospects. Curr Opin Biotechnol 19:210–217
Rezaul K, Wu LF, Mayya V, Hwang SI, Han D (2005) A systematic characterization of mitochondrial proteome from human T leukemia cells. Mol Cell Proteomics 4:169–181
Bendtsen JD, Nielsen H, von Heijne G, Brunak S (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340:783–795
Nielsen H, Engelbrecht J, Brunak S, von Heijne G (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng 10:1–6
van der Does C, Nouwen N, Driessen AJM (2003) The Sec translocase. In: Oudega B (ed) Protein secretion pathways in bacteria. Kluwer Academic Publishers, Boston, p 296
Bendtsen JD, Nielsen H, Widdick D, Palmer T, Brunak S (2005) Prediction of twin-arginine signal peptides. BMC Bioinform 6:167
Muesch A, Hartmann E, Rohde K, Rubartelli A, Sitia R, Rapoport TA (1990) A novel pathway for secretory proteins. Trends Biochem Sci 15:86–88
Rubartelli A, Cozzolino F, Talio M, Sitia R (1990) A novel secretory pathway for interleukin-1-beta, a protein lacking a signal sequence. EMBO J 9:1503–1510
Harth G, Clemens DL, Horwitz MA (1994) Glutamine synthetase of Mycobacterium tuberculosis: extracellular release and characterization of its enzymatic activity. PNAS 91:9342–9346
Harth G, Horwitz MA (1997) Expression and efficient export of enzymatically active Mycobacterium tuberculosis glutamine synthetase in Mycobacterium smegmatis and evidence that the information for export is contained within the protein. J Biol Chem 272:22728–22735
Bendtsen JD, Kiemer L, Fausboll A, Brunak S (2005) Non-classical protein secretion in bacteria. BMC Microbiol 5:58
Krogh A, Larsson B, von Heijne G, Sonnhammer ELL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580
Kall L, Krogh A, Sonnhammer ELL (2004) A combined transmembrane topology and signal peptide prediction method. J Mol Biol 338:1027–1036
Kall L, Krogh A, Sonnhammer ELL (2007) Advantages of combined transmembrane topology and signal peptide prediction—the Phobius web server. Nucleic Acids Res 35:W429–W432
van de Werken HJG, Verhaart MRA, VanFossen AL, Willquist K, Lewis DL, Nichols JD, Goorissen HP, Mongodin EF, Nelson KE, van Niel EWJ, Stams AJM, Ward DE, de Vos WM, van der Oost J, Kelly RM, Kengen SWM (2008) Hydrogenomics of the extremely thermophilic bacterium Caldicellulosiruptor saccharolyticus. Appl Environ Microbiol 74:6720–6729
Pierce Technical Resource (2004) TR00490. Pierce Technical Resource, Rockford, IL., USA
Jiang L, He L, Fountoulakis M (2004) Comparison of protein precipitation methods for sample preparation prior to proteomic analysis. J Chromatogr A 1023:317–320
Sauve DM, Ho DT, Roberge M (1995) Concentration of dilute protein for gel-electrophoresis. Anal Biochem 226:382–383
Winston RL, Fitzgerald MC (1998) Concentration and desalting of protein samples for mass spectrometry analysis. Anal Biochem 262:83–85
Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1:2856–2860
Andrews GL, Shuford CM, Burnett JC, Hawkridge AM, Muddiman DC (2009) Coupling of a vented column with splitless nanoRPLC-ESI-MS for the improved separation and detection of brain natriuretic peptide-32 and its proteolytic peptides. J Chromatogr B 877:948–954
Perkins DN, Pappin DJC, Creasy DM, Cottrell JS (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551–3567
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate—a practical and powerful approach to multiple testing. J R Stat Soc Ser B Stat Methodol 57:289–300
Weatherly DB, Atwood JA, Minning TA, Cavola C, Tarleton RL, Orlando R (2005) A heuristic method for assigning a false-discovery rate for protein identifications from mascot database search results. Mol Cell Proteomics 4:762–772
Zakowski J (1931) On the purification of soya-urease through precipitation with acetone and carbonic acids. Hoppe-Seyler's Z. Physiol Chem 202:67–82
Thongboonkerd V, McLeish KR, Arthur JM, Klein JB (2002) Proteomic analysis of normal human urinary proteins isolated by acetone precipitation or ultracentrifugation. J Am Soc Nephrol 13:119A–119A
Collier TS, Hawkridge AM, Georgianna DR, Payne GA, Muddiman DC (2008) Top-down identification and quantification of stable isotope labeled proteins from Aspergillus flavus using online nano-flow reversed-phase liquid chromatography coupled to a LTQ-FTICR mass spectrometer. Anal Chem 80:4994–5001
Sahyun M, Alsberg CL (1930) On rabbit liver glycogen and its preparation. J Biol Chem 89:33–39
Chevallet M, Diemer H, Van Dorssealer A, Villiers C, Rabilloud T (2007) Toward a better analysis of secreted proteins: the example of the myeloid cells secretome. Proteomics 7:1757–1770
Palmer JW, Gerlough TD (1940) A simple method for preparing antigenic substances from the typhoid bacillus. Science 92:155–156
Marusyk R, Sergeant A (1980) A simple method for dialysis of small-volume samples. Anal Biochem 105:403–404
Bergen HR, Vasmatzis G, Cliby WA, Johnson KL, Oberg AL, Muddiman DC (2003) Discovery of ovarian cancer biomarkers in serum using NanoLC electrospray ionization TOF and FT-ICR mass spectrometry. Dis Markers 19:239–249
Johnson KL, Mason CJ, Muddiman DC, Eckel JE (2004) Analysis of the low molecular weight fraction of serum by LC-dual ESI-FT-ICR mass spectrometry: precision of retention time, mass, and ion abundance. Anal Chem 76:5097–5103
Basrai MA, Hieter P, Boeke JD (1997) Small open reading frames: beautiful needles in the haystack. Genome Res 7:768–771
Gough J, Karplus K, Hughey R, Chothia C (2001) Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J Mol Biol 313:903–919
Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B (2007) Quantitative mass spectrometry in proteomics: a critical review. Anal Bioanal Chem 389:1017–1031
Gao J, Friedrichs MS, Dongre AR, Opiteck GJ (2005) Guidelines for the routine application of the peptide hits technique. J Am Soc Mass Spectrom 16:1231–1238
Liu HB, Sadygov RG, Yates JR (2004) A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem 76:4193–4201
Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, Resing KA, Ahn NG (2005) Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol Cell Proteomics 4:1487–1502
Zybailov B, Coleman MK, Florens L, Washburn MP (2005) Correlation of relative abundance ratios derived from peptide ion chromatograms and spectrum counting for quantitative proteomic analysis using stable isotope labeling. Anal Chem 77:6218–6224
Zybailov B, Mosley AL, Sardiu ME, Coleman MK, Florens L, Washburn MP (2006) Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J Proteome Res 5:2339–2347
Lynd LR, Weimer PJ, van Zyl WH, Pretorius IS (2002) Microbial cellulose utilization: fundamentals and biotechnology. Microbiol Mol Biol Rev 66:506–577
Lynd LR, Elander RT, Wyman CE (1996) Likely features and costs of mature biomass ethanol technology. Appl Biochem Biotechnol 57–8:741–761
Acknowledgements
We would like to acknowledge the financial support of the National Institutes of Health (Grant 5T32GM00-8776-08), which supports GLA in the North Carolina State University Molecular Biotechnology Training Program, and the W. M. Keck Foundation. RMK acknowledges support from the US National Science Foundation (CBT0617272) and the Bioenergy Science Center (BESC), a US DOE Bioenergy Research Center supported by the Office of Biological and Environmental Research. DLL acknowledges support from a US Department of Education GAANN Fellowship.
Author information
Authors and Affiliations
Corresponding author
Additional information
An erratum to this article can be found at http://dx.doi.org/10.1007/s00216-010-4102-0
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 551 kb)
Rights and permissions
About this article
Cite this article
Muddiman, D., Andrews, G., Lewis, D. et al. Part I: characterization of the extracellular proteome of the extreme thermophile Caldicellulosiruptor saccharolyticus by GeLC-MS2 . Anal Bioanal Chem 398, 377–389 (2010). https://doi.org/10.1007/s00216-010-3955-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-010-3955-6