Abstract
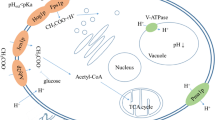
Extracellular conditions determine the taste of fermented foods by affecting metabolite formation by the micro-organisms involved. To identify targets for improvement of metabolite formation in food fermentation processes, automated high-throughput screening and cDNA microarray approaches were applied. Saccharomyces cerevisiae was cultivated in 96-well microtiter plates, and the effects of salt concentration and pH on the growth and synthesis of the fusel alcohol-flavoured substance, 3-methyl-1-butanol, was evaluated. Optimal fermentation conditions for 3-methyl-1-butanol concentration were found at pH 3.0 and 0% NaCl. To identify genes encoding enzymes with major influence on product formation, a genome-wide gene expression analysis was carried out with S. cerevisiae cells grown at pH 3.0 (optimal for 3-methyl-1-butanol formation) and pH 5.0 (yeast cultivated under standard conditions). A subset of 747 genes was significantly induced or repressed when the pH was changed from pH 5.0 to 3.0. Expression of seven genes related to the 3-methyl-1-butanol pathway, i.e. LAT1, PDX1, THI3, ALD4, ILV3, ILV5 and LEU4, strongly changed in response to this switch in pH of the growth medium. In addition, genes involved in NAD metabolism, i.e. BNA2, BNA3, BNA4 and BNA6, or those involved in the TCA cycle and glutamate metabolism, i.e. MEU1, CIT1, CIT2, KDG1 and KDG2, displayed significant changes in expression. The results indicate that this is a rapid and valuable approach for identification of interesting target genes for improvement of yeast strains used in industrial processes.






Similar content being viewed by others
References
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25:25–29
Bisson L (1999) Stuck and sluggish fermentations. Am J Enol Vitic 50:107–119
Boer VM, de Winde JH, Pronk JT, Piper MD (2003) The genome-wide trascriptional responses of Saccharomyces cerevisiae grown on glucose in aerobic chemostat cultures limited for carbon, nitrogen, phosporus, or sulfur. J Biol Chem 278:3265–3274
Colantuoni C, Henry G, Zeger S, Pevsner J (2002) SNOMAD (Standardization and NOrmalization of MicroArray Data): web-accessible gene expression data analysis. Bioinformatics 18:1540–1541
De Boer M (2000) Fermentative production of flavours in yeast. Thesis, Mol Biol Vrije Universiteit Amsterdam
De Boer M, Bebelman JP, Goncalves PM, Matt J, Van Heerikhuizen H, Planta RJ (1998) Regulation of expression of the amino acid transporter gene BAP3 in Saccharomyces cerevisiae. Mol Microbiol 30:603–613
Dickinson JR, Lanterman MM, Danner DJ, Pearson BM, Sanz P, Harrison SJ, Hewlins MJ (1997) A 13C nuclear magnetic resonance investigation of the metabolism of leucine to isoamyl alcohol in Saccharomyces cerevisiae. J Biol Chem 272:26871–26878
Dickinson JR, Salgado LE, Hewlins MJ (2003) The catabolism of amino acids to long chain and complex alcohols in Saccharomyces cerevisiae. J Biol Chem 278:8028–8034
Eden A, Van Nedervelde L, Drukker M, Benvenisty N, Debourg A (2001) Involvement of branched-chain amino acid aminotransferases in the production of fusel alcohols during fermentation in yeast. Appl Microbiol Biotechnol 55:296–300
Ehrlich F (1904) Uber das natürliche isomere des leucins. Dtsch Chem Ges 37:1809–1840
Forsberg H, Gilstring CF, Zargari A, Martinez P, Ljungdahl PO (2001) The role of the yeast plasma membrane SPS nutrient sensor in the metabolic response to extracellular amino acids. Mol Microbiol 42:215–228
Gasch AP, Werner-Washburne M (2002) The genomics of yeast responses to environmental stress and starvation. Funct Integr Genomics 2:181–192
Grauslund M, Didion T, Kielland-Bradt MC, Anderson HA (1995) BAP2, a gene encoding a permease for branched-chain amino acids in Saccharomyces cerevisiae. Biochim Biophys Acta 1269:275–280
Hammond JRM (1993) The yeasts, vol 5: yeast technology. Academic, London, pp 8–67
Jansen M, Veurink JH, Euverink GJ, Dijkhuizen L (2003) Growth of the salt-tolerant yeast Zygosaccharomyces rouxii in microtiter plates: effects of NaCl, pH and temperature on growth and fusel alcohol production from branched-chain amino acids. FEMS Yeast Res 3:313–318
Jauniaux JC, Grenson M (1990) GAP1, the general amino acid permease gene of Saccharomyces cerevisiae. Nucleotide sequence, protien similarily with the other bakers yeast amino acid permeases, and nitrogen catabolite repression. Eur J Biochem 190:39–44
Jia MH, Larossa RA, Lee JM, Rafalski A, Derose E, Gonye G, Xue Z (2000) Global expression profiling of yeast treated with an inhibitor of amino acid biosynthesis, sulfometuron methyl. Physiol Genomics 3:83–92
Kanehisa M (1997) A database for post-genome analysis. Trends Genet 13:375–376
Kanehisa M, Goto S (2000) KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30
Kispal G, Steiner H, Court DA, Rolinski B, Lill R (1996) Mitochondrial and cytosolic branched-chain amino acid transaminases from yeast, homologs of the myc oncogene-regulated Eca39 protein. J Biol Chem 271:24458–24464
Kodama Y, Omura F, Ashikari T (2001) Isolation and characterization of a gene specific to lager brewing yeast that encodes a branched-chain amino acid permease. Appl Environ Microbiol 67:3455–3462
Natarajan K, Meyer MR, Jackson BM, Slade D, Roberts C, Hinnebusch AG, Marton MJ (2001) Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol Cell Biol 21:4347–4368
Regenberg B, during-Olsen L, Keilland-Brandt MC, Holmberg S (1999) Substrate specificity and gene expression of the amino-acid permeases in Saccharomuces cerevisiae. Curr Genet 36:317 328
Ribéreau-Gayon P, Dubourdieu D, Donèche B, Lonvaud A (2000) Handbook of enology, vol 1. Wiley, New York
Schoondermark-Stolk SA, ter Schure EG, Verrips CT, Verkleij AJ, Boonstra J (2002) Identification of salt-induced genes of Zygosaccharomyces rouxii by using Saccharomyces cerevisiae GeneFilters. FEMS Yeast Res 2:525–532
Schoondermark-Stolk S, Tabernero M, Chapman J, Verrips T, Verkleij A, Boonstra J (2005) Bat2p is essential in Saccharomyces cerevisiae for fusel alcohol production on the non-fermentable carbon source ethanol. FEMS Yeast Res 5:757–766
Schreve J, Garrett JM (1997) The branched-chain amino acid permease gene of Saccharomyces cerevisiae, BAP2, encodes the high-affinity leucine permease (S1). Yeast 13:435–439
Stoops JK, Cheng RH, Yazdi MA, Maeng CY, Schroeter JP, Klueppelberg U, Kolodziej SJ, Baker TS, Reed LJ (1997) On the unique structural organization of the Saccharomyces cerevisiae pyruvate dehydrogenase complex. J Biol Chem 272:5757–5764
ter Schure EG, Flikweert MT, van Dijken JP, Pronk JT, Verrips CT (1998) Pyruvate decarboxylase catalyzes decarboxylation of branched-chain 2-oxo acids but is not essential for fusel alcohol production by Saccharomyces cerevisiae. Appl Environ Microbiol 64:1303–1307
Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 98:5116–5121
Verwaal R, Paalman JW, Hogenkamp A, Verkleij AJ, Verrips CT, Boonstra J (2002) HXT5 expression is determined by growth rates in Saccharomyces cerevisiae. Yeast 19:1029–1038
Volschenk H, Viljoen-Bloom M, Subden RE, van Vuuren HJ (2001) Malo-ethanolic fermentation in grape must by recombinant strains of Saccharomyces cerevisiae. Yeast 18:963–970
Weiss A, Delproposto J, Giroux CN (2004) High-throughput phenotypic profiling of gene–environment interactions by quantitative growth curve analysis in Saccharomyces cerevisiae. Anal Biochem 327:23–34
Yoshimoto H, Fukushige T, Yonezawa T, Sone H (2002) Genetic and physiological analysis of branched-chain alcohols and isoamyl acetate production in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 59:501–508
Acknowledgements
We thank Dr. Victor Winter (Utrecht School of Applied Science, Universiteit Utrecht, The Netherlands) for his assistance in the preparation of this manuscript. This research was supported by Senter Research Grant 99028 and sponsored by Unilever Research Vlaardingen.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Schoondermark-Stolk, S.A., Jansen, M., Veurink, J.H. et al. Rapid identification of target genes for 3-methyl-1-butanol production in Saccharomyces cerevisiae . Appl Microbiol Biotechnol 70, 237–246 (2006). https://doi.org/10.1007/s00253-005-0070-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-005-0070-2