Abstract
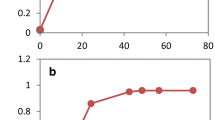
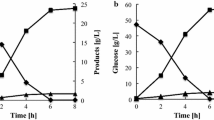
Engineering yeast to be more tolerant to fermentation inhibitors, furfural and 5-hydroxymethylfurfural (HMF), will lead to more efficient lignocellulose to ethanol bioconversion. To identify target genes involved in furfural tolerance, a Saccharomyces cerevisiae gene disruption library was screened for mutants with growth deficiencies in the presence of furfural. It was hypothesized that overexpression of these genes would provide a growth benefit in the presence of furfural. Sixty two mutants were identified whose corresponding genes function in a wide spectrum of physiological pathways, suggesting that furfural tolerance is a complex process. We focused on four mutants, zwf1, gnd1, rpe1, and tkl1, which represent genes encoding pentose phosphate pathway (PPP) enzymes. At various concentrations of furfural and HMF, a clear association with higher sensitivity to these inhibitors was demonstrated in these mutants. PPP mutants were inefficient at reducing furfural to the less toxic furfuryl alcohol, which we propose is a result of an overall decreased abundance of reducing equivalents or to NADPH's role in stress tolerance. Overexpression of ZWF1 in S. cerevisiae allowed growth at furfural concentrations that are normally toxic. These results demonstrate a strong relationship between PPP genes and furfural tolerance and provide additional putative target genes involved in furfural tolerance.



Similar content being viewed by others
References
Banerjee N, Bhatnagar R, Viswanathan L (1981) Inhibition of glycolysis by furfural in Saccharomyces cerevisiae. Eur J Appl Microbiol Biotechnol 11:226–228
Bothast RJ, Saha BC (1997) Ethanol production from agricultural biomass substrates. Adv Appl Microbiol 44:261–286
Carmel-Harel O, Storz G (2000) Roles of the glutathione- and thioredoxin-dependent reduction systems in the Escherichia coli and Saccharomyces cerevisiae responses to oxidative stress. Annu Rev Microbiol 54:439–461
Diaz de Villegas M, Villa P, Guerra M, Rodriguez E, Redondo D, Martinez A (1992) Conversion of furfural in furfuryl alcohol by Saccharomyces cerevisiae. Acta Biotechnol 12:351–354
Flores CL, Rodriguez C, Petit T, Gancedo C (2000) Carbohydrate and energy-yielding metabolism in non-conventional yeasts. FEMS Microbiol Rev 24:507–529
Gupta GD, Misra A, Agarwal DK (1991) Inhalation toxicity of furfural vapours: an assessment of biochemical response in rat lungs. J Appl Toxicol 11:343–347
Guthrie C, Fink GR (eds) (2002a) Methods in enzymology: guide to yeast genetics and molecular and cell biology, vol. 350 Part B. Academic, San Diego
Guthrie C, Fink GR (eds) (2002b) Methods in enzymology: guide to yeast genetics and molecular and cell biology, vol. 351 Part C. Academic, San Diego
Horvath IS, Taherzadeh MJ, Niklasson C, Liden G (2001) Effects of furfural on anaerobic continuous cultivation of Saccharomyces cerevisiae. Biotechnol Bioeng 75:540–549
Janzowski C, Glaab V, Samimi E, Schlatter J, Eisenbrand G (2000) 5-Hydroxymethylfurfural: assessment of mutagenicity, DNA-damaging potential and reactivity towards cellular glutathione. Food Chem Toxicol 38:801–809
Jeppsson M, Johansson B, Jensen PR, Hahn-Hagerdal B, Gorwa-Grauslund MF (2003) The level of glucose-6-phosphate dehydrogenase activity strongly influences xylose fermentation and inhibitor sensitivity in recombinant Saccharomyces cerevisiae strains. Yeast 20:1263–1272
Juhnke H, Krems B, Kotter P, Entian KD (1996) Mutants that show increased sensitivity to hydrogen peroxide reveal an important role for the pentose phosphate pathway in protection of yeast against oxidative stress. Mol Gen Genet 252:456–464
Karhumaa K, Hahn-Hagerdal B, Gorwa-Grauslund MF (2005) Investigation of limiting metabolic steps in the utilization of xylose by recombinant Saccharomyces cerevisiae using metabolic engineering. Yeast 22:359–368
Krems B, Charizanis C, Entian KD (1995) Mutants of Saccharomyces cerevisiae sensitive to oxidative and osmotic stress. Curr Genet 27:427–434
Kuyper M, Hartog MM, Toirkens MJ, Almering MJ, Winkler AA, van Dijken JP, Pronk JT (2005) Metabolic engineering of a xylose-isomerase-expressing Saccharomyces cerevisiae strain for rapid anaerobic xylose fermentation. FEMS Yeast Res 5:399–409
Larroy C, Fernandez MR, Gonzalez E, Pares X, Biosca JA (2002) Characterization of the Saccharomyces cerevisiae YMR318C (ADH6) gene product as a broad specificity NADPH-dependent alcohol dehydrogenase: relevance in aldehyde reduction. Biochem J 361:163–172
Larsson S, Palmqvist E, Hahn-Hagerdal B, Tengborg C, Stenberg K, Zacchi G, Nilvebrant N-O (1999) The generation of inhibitors during dilute acid hydrolysis of softwood. Enzyme Microb Technol 24:151–159
Liu ZL, Slininger PJ, Dien BS, Berhow MA, Kurtzman CP, Gorsich SW (2004) Adaptive response of yeasts to furfural and 5-hydroxymethylfurfural and new chemical evidence for HMF conversion to 2,5-bis-hydroxymethylfuran. J Ind Microbiol Biotech 31:345–352
Liu ZL, Slininger PJ, Gorsich SW (2005) Enhanced biotransformation of furfural and hydroxymethylfurfural by newly developed ethanologenic yeast strains. Appl Biochem Biotechnol 121–124:451–460
Lynd L (1996) Overview and evaluation of fuel ethanol from cellulosic biomass: technology, economics, the environment, and policy. Annu Rev Energy Environ 21:403–465
Maciel de Mancilha I, Karim MN (2003) Evaluation of ion exchange resins for removal of inhibitory compounds from corn stover hydrolyzate for xylitol fermentation. Biotechnol Prog 19:1837–1841
Modig T, Liden G, Taherzadeh MJ (2002) Inhibition effects of furfural on alcohol dehydrogenase, aldehyde dehydrogenase and pyruvate dehydrogenase. Biochem J 363:769–776
Mollapour M, Fong D, Balakrishnan K, Harris N, Thompson S, Schuller C, Kuchler K, Piper PW (2004) Screening the yeast deletant mutant collection for hypersensitivity and hyper-resistance to sorbate, a weak organic acid food preservative. Yeast 21:927–946
Nogae I, Johnston M (1990) Isolation and characterization of the ZWF1 gene of Saccharomyces cerevisiae, encoding glucose-6-phosphate dehydrogenase. Gene 96:161–169
Olsson L, Hahn-Hagerbal B (1996) Fermentation of lignocellulosic hydrolysates for ethanol production. Enzyme Microb Technol 18:312–331
Palmqvist E, Hahn-Hagerdal B (2000) Fermentation of lignocellulosic hydrolysates. II: inhibitors and mechanisms of inhibition. Bioresour Technol 74:25–33
Palmqvist E, Almeida JS, Hahn-Hagerdal B (1999) Influence of furfural on anaerobic glycolytic kinetics of Saccharomyces cerevisiae in batch culture. Biotechnol Bioeng 62:447–454
Saha BC (2003) Hemicellulose bioconversion. J Ind Microbiol Biotech 30:279–291
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, New York
Sanchez BBJ (1988) Effects of furfural and 5-hydroxymethylfurfural on the fermentation of Saccharomyces cerevisiae and biomass production from Candida guilliermondii. Enzyme Microb Technol 10:315–318
Schneider R, Brors B, Burger F, Camrath S, Weiss H (1997) Two genes of the putative mitochondrial fatty acid synthase in the genome of Saccharomyces cerevisiae. Curr Genet 32:384–388
Slekar KH, Kosman DJ, Culotta VC (1996) The yeast copper/zinc superoxide dismutase and the pentose phosphate pathway play overlapping roles in oxidative stress protection. J Biol Chem 271:28831–28836
Swan TM, Watson K (1999) Stress tolerance in a yeast lipid mutant: membrane lipids influence tolerance to heat and ethanol independently of heat shock proteins and trehalose. Can J Microbiol 45:472–479
Taherzadeh MJ, Gustafsson L, Niklasson C, Liden G (1999) Conversion of furfural in aerobic and anaerobic batch fermentation of glucose by Saccharomyces cerevisiae. J Biosci Bioeng 87:169–174
Taherzadeh MJ, Gustafsson L, Niklasson C, Liden G (2000a) Inhibition effects of furfural on aerobic batch cultivation of Saccharomyces cerevisiae growing on ethanol and/or acetic acid. J Biosci Bioeng 90:374–380
Taherzadeh MJ, Gustafsson L, Niklasson C, Liden G (2000b) Physiological effects of 5-hydroxymethylfurfural on Saccharomyces cerevisiae. Appl Microbiol Biotechnol 53:701–708
Thorpe GW, Fong CS, Alic N, Higgins VJ, Dawes IW (2004) Cells have distinct mechanisms to maintain protection against different reactive oxygen species: oxidative-stress-response genes. Proc Natl Acad Sci U S A 101:6564–6569
Villa GP, Bartroli R, Lopez R, Guerra M, Enrique M, Penas M, Rodriguez E, Redondo D, Iglesias I, Diaz M (1992) Microbial transformation of furfural to furfuryl alcohol by Saccharomyces cerevisiae. Acta Biotechnol 12:509–512
Wahlbom CF, Hahn-Hagerdal B (2002) Furfural, 5-hydroxymethyl furfural, and acetoin act as external electron acceptors during anaerobic fermentation of xylose in recombinant Saccharomyces cerevisiae. Biotechnol Bioeng 78:172–178
Wahlbom CF, Cordero Otero RR, van Zyl WH, Hahn-Hagerdal B, Jonsson LJ (2003) Molecular analysis of a Saccharomyces cerevisiae mutant with improved ability to utilize xylose shows enhanced expression of proteins involved in transport, initial xylose metabolism, and the pentose phosphate pathway. Appl Environ Microbiol 69:740–746
Wheals AE, Basso LC, Alves DM, Amorim HV (1999) Fuel ethanol after 25 years. Trends Biotechnol 17:482–487
Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Dow SW, El Bakkoury M, Foury F, Friend SH, Gentalen E, Giaever G, Hegemann JH, Jones T, Laub M, Liao H, Davis RW et al (1999) Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901–906
Zaldivar J, Nielsen J, Olsson L (2001) Fuel ethanol production from lignocellulose: a challenge for metabolic engineering and process integration. Appl Microbiol Biotechnol 56:17–34
Zaldivar J, Borges A, Johansson B, Smits HP, Villas-Boas SG, Nielsen J, Olsson L (2002) Fermentation performance and intracellular metabolite patterns in laboratory and industrial xylose-fermenting Saccharomyces cerevisiae. Appl Microbiol Biotechnol 59:436–442
Acknowledgements
Special thanks to Drs. Nancy Alexander and Cletus Kurtzman for helpful discussions and suggestions and to Nicole Maroon, Stephanie Gove, Elizabeth Kilduski, Sarah Frazer, and Patricia O'Bryan for expert technical assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture.
Rights and permissions
About this article
Cite this article
Gorsich, S.W., Dien, B.S., Nichols, N.N. et al. Tolerance to furfural-induced stress is associated with pentose phosphate pathway genes ZWF1, GND1, RPE1, and TKL1 in Saccharomyces cerevisiae . Appl Microbiol Biotechnol 71, 339–349 (2006). https://doi.org/10.1007/s00253-005-0142-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-005-0142-3