Abstract
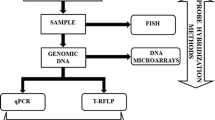
Since the application of molecular methods, culture-independent methods (CIMs) have been developed to study microbial communities from various environments. In the past 20 years, several methods based on the direct amplification and analyses of the small subunit ribosomal RNA gene have been developed to directly study environmental microorganisms. These methods include denaturing/temperature gradient gel electrophoresis, single-strand-conformation polymorphism, restriction fragment length polymorphism, terminal restriction fragment length polymorphism, and quantitative polymerase chain reaction (PCR). Similarly, non-PCR-based molecular techniques, such as microarray and fluorescence in situ hybridization have also been adopted. In recent years, several novel fields of investigation such as metagenomics, metatranscriptomics, metaproteomics, and single-cell genomics were developed, largely propelled by the innovation and application of next-generation sequencing methods. Several single-cell-based technologies such as Raman microspectroscopy and nano-scale secondary ion mass spectrometry are also increasingly used in the fields of microbial ecology and environmental microbiology. The application of these methods has revolutionized microbiology by allowing scientists to directly analyze natural microbial communities in situ, including their genes, transcripts, proteins, and metabolites and how their interactions impact their distribution patterns. In this review, we present an up-to-date review on different CIMs and their applications, our focuses are on the comparison of different CIMs and their application in the analyses of microbial diversities and communities.

Similar content being viewed by others
References
Amann RI, Ludwig W, Schleifer KH (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59:143–169
Andersson AF, Lindberg M, Jakobsson H, Backhed F, Nyren P, Engstrand L (2008) Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One 3:e2836
Armougom F, Henry M, Vialettes B, Raccah D, Raoult D (2009) Monitoring bacterial community of human gut microbiota reveals an increase in Lactobacillus in obese patients and methanogens in anorexic patients. PLoS One 4:e7125
Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI (2005) Host–bacterial mutualism in the human intestine. Science 307:1915–1920
Baik KS, Park SC, Kim EM, Bae KS, Ahn JH, Ka JO, Chun J, Seong CN (2008) Diversity of bacterial community in freshwater of Woopo wetland. J Microbiol 46:647–655
Behrens S, Lösekann T, Pett-Ridge J, Weber PK, Ng WO, Stevenson BS, Hutcheon ID, Relman DA, Spormann AM (2008) Linking microbial phylogeny to metabolic activity at the single-cell level by using enhanced element labeling-catalyzed reporter deposition fluorescence in situ hybridization (EL-FISH) and NanoSIMS. Appl Environ Microbiol 74:3143–3150
Benndorf D, Balcke GU, Harms H, von Bergen M (2007) Functional metaproteome analysis of protein extracts from contaminated soil and groundwater. ISME J 1:224–234
Besser TE, Cassirer EF, Potter KA, VanderSchalie J, Fischer A, Knowles DP, Herndon DR, Rurangirwa FR, Weiser GC, Srikumaran S (2008) Association of Mycoplasma ovipneumoniae infection with population-limiting respiratory disease in free-ranging Rocky Mountain bighorn sheep (Ovis canadensis canadensis). J Clin Microbiol 46:423–430
Bik EM, Long CD, Armitage GC, Loomer P, Emerson J, Mongodin EF, Nelson KE, Gill SR, Fraser-Liggett CM, Relman DA (2010) Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J 4:962–974
Bjorklof K, Karlsson S, Frostegard A, Jorgensen KS (2009) Presence of actinobacterial and fungal communities in clean and petroleum hydrocarbon contaminated subsurface soil. Open Microbiol J 3:75–86
Blainey PC, Mosier AC, Potanina A, Francis CA, Quake SR (2011) Genome of a low-salinity ammonia-oxidizing archaeon determined by single-cell and metagenomic analysis. PLoS One 6:e16626
Bodrossy L, Sessitsch A (2004) Oligonucleotide microarrays in microbial diagnostics. Curr Opin Microbiol 7:245–254
Bottari B, Ercolini D, Gatti M, Neviani E (2006) Application of FISH technology for microbiological analysis: current state and prospects. Appl Microbiol Biotechnol 73:485–494
Brehm-Stecher BF, Johnson EA (2004) Single-cell microbiology: tools, technologies, and applications. Microbiol Mol Biol Rev 68:538–539
Bruce T, Martinez IB, Maia Neto O, Vicente AC, Kruger RH, Thompson FL (2010) Bacterial community diversity in the Brazilian Atlantic forest soils. Microb Ecol 60:840–849
Chen Y, Wu L, Boden R, Hillebrand A, Kumaresan D, Moussard H, Baciu M, Lu Y, Colin Murrell J (2009) Life without light: microbial diversity and evidence of sulfur-and ammonium-based chemolithotrophy in Movile Cave. ISME J 3:1093–1104
Chitsaz H, Yee-Greenbaum JL, Tesler G, Lombardo MJ, Dupont CL, Badger JH, Novotny M, Rusch DB, Fraser LJ, Gormley NA, Schulz-Trieglaff O, Smith GP, Evers DJ, Pevzner PA, Lasken RS (2011) Efficient de novo assembly of single-cell bacterial genomes from short-read data sets. Nat Biotechnol 29:915–921
Connon SA, Giovannoni SJ (2002) High-throughput methods for culturing microorganisms in very-low-nutrient media yield diverse new marine isolates. Appl Environ Microbiol 68:3878–3885
Cruz-Martínez K, Suttle KB, Brodie EL, Power ME, Andersen GL, Banfield JF (2009) Despite strong seasonal responses, soil microbial consortia are more resilient to long-term changes in rainfall than overlying grassland. ISME J 3:738–744
Dekas AE, Orphan VJ (2011) Identification of diazotrophic microorganisms in marine sediment via fluorescence in situ hybridization coupled to nanoscale secondary ion mass spectrometry (FISH–NanoSIMS). Methods Enzymol 486:281–305
Delgado S, Arroyo R, Martin R, Rodriguez JM (2008) PCR-DGGE assessment of the bacterial diversity of breast milk in women with lactational infectious mastitis. BMC Infect Dis 8:51
Dethlefsen L, Relman DA (2011) Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci USA 108:4554–4561
Dethlefsen L, Huse S, Sogin ML, Relman DA (2008) The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol 6:e280
Di Giacomo M, Paolino M, Silvestro D, Vigliotta G, Imperi F, Visca P, Alifano P, Parente D (2007) Microbial community structure and dynamics of dark fire-cured tobacco fermentation. Appl Environ Microbiol 73:825–837
Diez B, Bauer K, Bergman B (2007) Epilithic cyanobacterial communities of a marine tropical beach rock (Heron Island, Great Barrier Reef): diversity and diazotrophy. Appl Environ Microbiol 73:3656–3668
Dunbar J, Ticknor LO, Kuske CR (2000) Assessment of microbial diversity in four southwestern United States soils by 16S rRNA gene terminal restriction fragment analysis. Appl Environ Microbiol 66:2943–2950
Engel AS, Meisinger DB, Porter ML, Payn RA, Schmid M, Stern LA, Schleifer KH, Lee NM (2010) Linking phylogenetic and functional diversity to nutrient spiraling in microbial mats from Lower Kane Cave (USA). ISME J 4:98–110
Fierer N, Liu Z, Rodriguez-Hernandez M, Knight R, Henn M, Hernandez MT (2008) Short-term temporal variability in airborne bacterial and fungal populations. Appl Environ Microbiol 74:200–207
Frias-Lopez J, Shi Y, Tyson GW, Coleman ML, Schuster SC, Chisholm SW, Delong EF (2008) Microbial community gene expression in ocean surface waters. Proc Natl Acad Sci USA 105:3805–3810
Fröhlich-Nowoisky J, Pickersgill DA, Després VR, Pöschl U (2009) High diversity of fungi in air particulate matter. Proc Natl Acad Sci USA 106:12814–12819
Goll J, Rusch DB, Tanenbaum DM, Thiagarajan M, Li K, Methé BA, Yooseph S (2010) METAREP: JCVI metagenomics reports—an open source tool for high-performance comparative metagenomics. Bioinformatics 26:2631–2632
Gugliandolo C, Lentini V, Maugeri TL (2010) Distribution and diversity of bacteria in a saline meromictic lake as determined by PCR-DGGE of 16S rRNA gene fragments. Curr Microbiol 62:159–166
He S, Kunin V, Haynes M, Martin HG, Ivanova N, Rohwer F, Hugenholtz P, McMahon KD (2010) Metatranscriptomic array analysis of” ‘Candidatus Accumulibacter phosphatis’-enriched enhanced biological phosphorus removal sludge. Environ Microbiol 12:1205–1217
Huang WE, Griffiths RI, Thompson IP, Bailey MJ, Whiteley AS (2004) Raman microscopic analysis of single microbial cells. Anal Chem 76:4452–4458
Huang WE, Ferguson A, Singer A, Lawson K, Thompson IP, Kalin RM, Larkin MJ, Bailey MJ, Whiteley AS (2009) Resolving genetic functions within microbial populations: in situ analyses using rRNA and mRNA stable isotope probing coupled with single-cell Raman-fluorescence in situ hybridization. Appl Environ Microbiol 75:234–241
Huang JW, Yang JK, Duan YQ, Gu W, Gong XW, Zhe W, Su C, Zhang KQ (2010a) Bacterial diversities on unaged and aging flue-cured tobacco leaves estimated by 16S rRNA sequence analysis. Appl Microbiol Biotechnol 88:553–562
Huang WE, Li M, Jarvis RM, Goodacre R, Banwart SA (2010b) Shining light on the microbial world the application of Raman microspectroscopy. Adv Appl Microbiol 70:153–186
Hutter G, Schlagenhauf U, Valenza G, Horn M, Burgemeister S, Claus H, Vogel U (2003) Molecular analysis of bacteria in periodontitis: evaluation of clone libraries, novel phylotypes and putative pathogens. Microbiology 149:67–75
Ishoey T, Woyke T, Stepanauskas R, Novotny M, Lasken RS (2008) Genomic sequencing of single microbial cells from environmental samples. Curr Opin Microbiol 11:198–204
Jones SE, Lennon JT (2010) Dormancy contributes to the maintenance of microbial diversity. Proc Natl Acad Sci USA 107:5881–5886
Kim YH, Kim IS, Moon EY, Park JS, Kim SJ, Lim JH, Park BT, Lee EJ (2011) High abundance and role of antifungal bacteria in compost-treated soils in a wildfire area. Microb Ecol 62:725–737
Koren O, Spor A, Felin J, Fak F, Stombaugh J, Tremaroli V, Behre CJ, Knight R, Fagerberg B, Ley RE, Backhed F (2011) Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc Natl Acad Sci USA 108:4592–4598
Kuypers MMM (2007) Microbiology: sizing up the uncultivated majority. Science 317:1510–1511
Kuypers MMM, Jorgensen BB (2007) The future of single-cell environmental microbiology. Environ Microbiol 9:6–7
Kvist T, Ahring BK, Lasken RS, Westermann P (2007) Specific single-cell isolation and genomic amplification of uncultured microorganisms. Appl Microbiol Biotechnol 74:926–935
Laguerre G, Allard MR, Revoy F, Amarger N (1994) Rapid identification of rhizobia by restriction-fragment-length-polymorphism analysis of PCR-amplified 16S ribosomal-RNA genes. Appl Environ Microbiol 60:56–63
Lasken RS (2007) Single-cell genomic sequencing using multiple displacement amplification. Curr Opin Microbiol 10:510–516
Lee DH, Zo YG, Kim SJ (1996) Nonradioactive method to study genetic profiles of natural bacterial communities by PCR single-strand-conformation polymorphism. Appl Environ Microbiol 62:3112–3120
Lemon KP, Klepac-Ceraj V, Schiffer HK, Brodie EL, Lynch SV, Kolter R (2010) Comparative analyses of the bacterial microbiota of the human nostril and oropharynx. MBio 1:e00129–10
Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI (2005) Obesity alters gut microbial ecology. Proc Natl Acad Sci USA 102:11070–11075
Ley RE, Turnbaugh PJ, Klein S, Gordon JI (2006) Microbial ecology: human gut microbes associated with obesity. Nature 444:1022–1023
Ling ZX, Liu X, Chen XY, Zhu HB, Nelson KE, Xia YX, Li LJ, Xiang CL (2011) Diversity of cervicovaginal microbiota associated with female lower genital tract infections. Microb Ecol 61:704–714
Löhr AJ, Laverman AM, Braster M, Van Straalen NM, Röling WFM (2006) Microbial communities in the world's largest acidic volcanic lake, Kawah Ijen in Indonesia, and in the Banyupahit River originating from it. Microb Ecol 52:609–618
Muyzer G (1999) DGGE/TGGE a method for identifying genes from natural ecosystems. Curr Opin Microbiol 2:317–322
Nocker A, Burr M, Camper AK (2007) Genotypic microbial community profiling: a critical technical review. Microb Ecol 54:276–289
Nowrousian M (2010) Next-generation sequencing techniques for eukaryotic microorganisms: sequencing-based solutions to biological problems. Eukaryot Cell 9:1300–1310
Oehler DZ, Robert F, Walter MR, Sugitani K, Meibom A, Mostefaoui S, Gibson EK (2010) Diversity in the Archean biosphere: new insights from NanoSIMS. Astrobiology 10:413–424
Pace NR, Stahl DA, Lane DJ, Olsen G (1985) Analyzing natural microbial populations by rRNA sequences. ASM News 51:4–12
Poretsky RS, Gifford S, Rinta-Kanto J, Vila-Costa M, Moran MA (2009) Analyzing gene expression from marine microbial communities using environmental transcriptomics. JoVE. 24. http://www.jove.com/index/Details.stp?ID=1086, doi: 10.3791/1086
Raghunathan A, Ferguson HR Jr, Bornarth CJ, Song W, Driscoll M, Lasken RS (2005) Genomic DNA amplification from a single bacterium. Appl Environ Microbiol 71:3342–3347
Rani A, Sharma A, Rajagopal R, Adak T, Bhatnagar RK (2009) Bacterial diversity analysis of larvae and adult midgut microflora using culture-dependent and culture-independent methods in lab-reared and field-collected Anopheles stephensi—an Asian malarial vector. BMC Microbiol 9:96
Rastogi G, Stetler LD, Peyton BM, Sani RK (2009) Molecular analysis of prokaryotic diversity in the deep subsurface of the former Homestake gold mine, South Dakota, USA. J Microbiol 47:371–384
Rastogi G, Osman S, Kukkadapu R, Engelhard M, Vaishampayan PA, Andersen GL, Sani RK (2010a) Microbial and mineralogical characterizations of soils collected from the deep biosphere of the former Homestake gold mine, South Dakota. Microb Ecol 59:1–12
Rastogi G, Osman S, Vaishampayan PA, Andersen GL, Stetler LD, Sani RK (2010b) Microbial diversity in uranium mining-impacted soils as revealed by high-density 16S microarray and clone library. Microb Ecol 59:94–108
Reynisson E, Lauzon H, Magnússon H, Jónsdóttir R, Ólafsdóttir G, Marteinsson V, Hreggviðsson GÓ (2009) Bacterial composition and succession during storage of North-Atlantic cod (Gadus morhua) at superchilled temperatures. BMC Microbiol 9:250
Rober M, Quirynen M, Haffajee AD, Schepers E, Teughels W (2008) Intra-oral microbial profiles of beagle dogs assessed by checkerboard DNA–DNA hybridization using human probes. Vet Microbiol 127:79–88
Roesch LF, Fulthorpe RR, Riva A, Casella G, Hadwin AK, Kent AD, Daroub SH, Camargo FA, Farmerie WG, Triplett EW (2007) Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J 1:283–290
Roth E, Miescher Schwenninger S, Hasler M, Eugster-Meier E, Lacroix C (2010) Population dynamics of two antilisterial cheese surface consortia revealed by temporal temperature gradient gel electrophoresis. BMC Microbiol 10:74
Sagaram US, DeAngelis KM, Trivedi P, Andersen GL, Lu SE, Wang N (2009) Bacterial diversity analysis of Huanglongbing pathogen-infected citrus, using PhyloChip arrays and 16S rRNA gene clone library sequencing. Appl Environ Microbiol 75:1566–1574
Scanlan PD, Marchesi JR (2008) Micro-eukaryotic diversity of the human distal gut microbiota: qualitative assessment using culture-dependent and -independent analysis of faeces. ISME J 2:1183–1193
Schneider T, Riedel K (2010) Environmental proteomics: analysis of structure and function of microbial communities. Proteomics 10:785–798
Shravage BV, Dayananda KM, Patole MS, Shouche YS (2007) Molecular microbial diversity of a soil sample and detection of ammonia oxidizers from Cape Evans, Mcmurdo Dry Valley, Antarctica. Microbiol Res 162:15–25
Siala M, Gdoura R, Fourati H, Rihl M, Jaulhac B, Younes M, Sibilia J, Baklouti S, Bargaoui N, Sellami S, Sghir A, Hammami A (2009) Broad-range PCR, cloning and sequencing of the full 16S rRNA gene for detection of bacterial DNA in synovial fluid samples of Tunisian patients with reactive and undifferentiated arthritis. Arthritis Res Ther 11:R102
Simon C, Daniel R (2011) Metagenomic analyses: past and future trends. Appl Environ Microbiol 77:1153–1161
Smith JJ, Tow LA, Stafford W, Cary C, Cowan DA (2006) Bacterial diversity in three different Antarctic Cold Desert mineral soils. Microb Ecol 51:413–421
Sorek R, Cossart P (2010) Prokaryotic transcriptomics: a new view on regulation, physiology and pathogenicity. Nat Rev Genet 11:9–16
Sowell SM, Wilhelm LJ, Norbeck AD, Lipton MS, Nicora CD, Barofsky DF, Carlson CA, Smith RD, Giovanonni SJ (2009) Transport functions dominate the SAR11 metaproteome at low-nutrient extremes in the Sargasso Sea. ISME J 3:93–105
Su C, Gu W, Zhe W, Zhang KQ, Duan YQ, Yang JK (2011) Diversity and phylogeny of bacteria on Zimbabwe tobacco leaves estimated by 16S rRNA sequence analysis. Appl Microbiol Biotechnol 92:1033–1044
Szekely BA, Singh J, Marsh TL, Hagedorn C, Werre SR, Kaur T (2010) Fecal bacterial diversity of human-habituated wild chimpanzees (Pan troglodytes schweinfurthii) at Mahale Mountains National Park, Western Tanzania. Am J Primatol 72:566–574
Takai K, Horikoshi K (2000) Rapid detection and quantification of members of the archaeal community by quantitative PCR using fluorogenic probes. Appl Environ Microbiol 66:5066–5072
Thompson JR, Marcelino LA, Polz MF (2002) Heteroduplexes in mixed-template amplifications: formation, consequence and elimination by ‘reconditioning PCR’. Nucleic Acids Res 30:2083–2088
Tringe SG, Zhang T, Liu X, Yu Y, Lee WH, Yap J, Yao F, Suan ST, Ing SK, Haynes M, Rohwer F, Wei CL, Tan P, Bristow J, Rubin EM, Ruan Y (2008) The airborne metagenome in an indoor urban environment. PLoS One 3:e1862
Urich T, Lanzén A, Qi J, Huson DH, Schleper C, Schuster SC (2008) Simultaneous assessment of soil microbial community structure and function through analysis of the meta-transcriptome. PLoS One 3:e2527
Verberkmoes NC, Russell AL, Shah M, Yu Y, Lee WH, Yap J, Yao F, Suan ST, Ing SK, Haynes M, Rohwer F, Wei CL, Tan P, Bristow J, Rubin EM, Ruan Y (2009) Shotgun metaproteomics of the human distal gut microbiota. ISME J 3:179–189
Vianna ME, Conrads G, Gomes BP, Horz HP (2009) T-RFLP-based mcrA gene analysis of methanogenic archaea in association with oral infections and evidence of a novel Methanobrevibacter phylotype. Oral Microbiol Immunol 24:417–422
Vladar P, Rusznyak A, Marialigeti K, Borsodi AK (2008) Diversity of sulfate-reducing bacteria inhabiting the rhizosphere of Phragmites australis in Lake Velencei (Hungary) revealed by a combined cultivation-based and molecular approach. Microb Ecol 56:64–75
von Mering C, Hugenholtz P, Raes J, Tringe SG, Doerks T, Jensen LJ, Ward N, Bork P (2007) Quantitative phylogenetic assessment of microbial communities in diverse environments. Science 315:1126–1130
Wang Y, Hoenig JD, Malin KJ, Qamar S, Petrof EO, Sun J, Antonopoulos DA, Chang EB, Claud EC (2009) 16S rRNA gene-based analysis of fecal microbiota from preterm infants with and without necrotizing enterocolitis. ISME J 3:944–954
Wasmund K, Kurtböke DI, Burns KA, Bourne DG (2009) Microbial diversity in sediments associated with a shallow methane seep in the tropical Timor Sea of Australia reveals a novel aerobic methanotroph diversity. FEMS Microbiol Ecol 68:142–151
Wilmes P, Bond PL (2004) The application of two-dimensional polyacrylamide gel electrophoresis and downstream analyses to a mixed community of prokaryotic microorganisms. Environ Microbiol 6:911–920
Wilmes P, Wexler M, Bond PL (2008) Metaproteomics provides functional insight into activated sludge wastewater treatment. PLoS One 3:e1778
Woyke T, Tighe D, Mavromatis K, Clum A, Copeland A, Schackwitz W, Lapidus A, Wu D, McCutcheon JP, McDonald BR (2010) One bacterial cell, one complete genome. PLoS One 5:e10314
Xie CG, Li YQ (2002) Raman spectra and optical trapping of highly refractive and nontransparent particles. Appl Phys Lett 81:951–953
Xie C, Mace J, Dinno MA, Li YQ, Tang W, Newton RJ, Gemperline PJ (2005) Identification of single bacterial cells in aqueous solution using conflocal laser tweezers Raman spectroscopy. Anal Chem 77:4390–4397
Xu JP (2006) Microbial ecology in the age of genomics and metagenomics: concepts, tools and recent advances. Mol Ecol 15:1713–1731
Xu B, Huang ZX, Wang XY, Gao RC, Tang XH, Mu YL, Yang YJ, Shi H, Zhu LD (2010) Phylogenetic analysis of the fecal flora of the wild pygmy loris. Am J Primatol 72:699–706
Yuan J, Zeng BH, Niu R, Tang H, Li WX, Zhang ZX, Wei H (2011) The development and stability of the genus Bacteriodes from human gut microbiota in HFA mice model. Curr Microbiol 62:1107–1112
Zhang W, Wang H, Zhang R, Yu XZ, Qian PY, Wong MH (2010) Bacterial communities in PAH contaminated soils at an electronic-waste processing center in China. Ecotoxicology 19:96–104
Acknowledgements
We are grateful to Prof. Jianping Xu (McMaster University, Canada) and Dr. Baoyu Tian (Yale University, USA) for their valuable comments and critical discussions. This work was supported by the National Natural Science Foundation of China (approval nos. U1036602 and 31060012), the West Light Foundation of the Chinese Academy of Sciences (to Jinkui Yang), and by projects from the Department of Science and Technology of Yunnan Province (approval nos. 2006GG22 and 2009CI052) and the Yunnan Branch of China Tobacco Industrial Corporation (approval no. 2010yn16).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Su, C., Lei, L., Duan, Y. et al. Culture-independent methods for studying environmental microorganisms: methods, application, and perspective. Appl Microbiol Biotechnol 93, 993–1003 (2012). https://doi.org/10.1007/s00253-011-3800-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-011-3800-7