Abstract
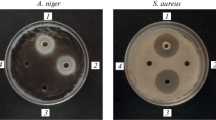
Lipopeptides represent a unique class of bioactive microbial secondary metabolites, and iturin A shows attractive antibiotic properties among them. This study compares three methods, such as yeast/fungal growth inhibition assay, quantitative high-performance liquid chromatography (HPLC) and polymerase chain reaction (PCR) for identifying a number of Bacillus species that produce iturin A. We examined the feasibility of screening iturin A-producing Bacillus strains by PCR using specific primers for ituD and lpa-14 amplification. Twenty standard strains and 120 field-collected Bacillus spp. isolates were tested in this study. Four B. subtilis and one B. circulan strains from ATCC, and B. amyloliquefaciens B128, a known iturin A producer, exhibited positive results. Of the 120 field-collected isolates, 42 strains were positive. The potential of producing iturin A by these PCR-positive strains were then confirmed by conventional methods such as fungal growth inhibition assay and HPLC analysis. The consistency between results of PCR, HPLC, and fungal growth inhibition assay suggests that the PCR method could be used as an alternative tool for fast screening of iturin A-producing Bacillus strains from the environment. This is the first report of detecting iturin A production from B. circulans.

Similar content being viewed by others
References
De Lucca AJ, Walsh TJ (1999) Antifungal peptide: novel therapeutic compounds against emerging pathogens. Antimicrob Agents Chemother 43:1–11
Feignier C, Besson F, Michel G (1995) Studies on lipopeptide biosynthesis by Bacillus subtilis: isolation and characterization of iturin-, surfactin+ mutants. FEMS Microbiol Lett 127:11–15
Hiraoka H, Ano T, Shoda M (1992) Molecular cloning of a gene responsible for the biosynthesis of the lipopeptide antibiotics iturin and surfactin. J Ferment Bioeng 74:323–326
Hsieh FC, Li MC, Lin TC, Kao SS (2004) Rapid detection and characterization of surfactin-producing Bacillus subtilis and closely related species based on PCR. Curr Microbiol 49:186–191
Huang CC, Ano T, Shoda M (1993) Nucleotide sequence and characteristics of the gene, lpa-14, responsible for biosynthesis of the lipopeptide antibiotics iturin A and surfactin from Bacillus subtilis RB14. J Ferment Bioeng 76:445–450
Kunst F, Ogasawara N, Moszer I (1997) The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390:249–256
Latoud C, Peypoux F, Michel G (1987) Action of iturin A, an antifungal antibiotic from Bacillus subtilis, on the yeast Saccharomyces cerevisiae: modification of membrane permeability and lipid composition. J Antibiot 11:1588–1595
Maget-Dana R, Peypoux F (1994) Iturins, a special class of pore-forming lipopeptides: biological and physicochemical properties. Toxicology 87:151–174
Ohno A, Ano T, Shoda M (1993) Production of the antifungal peptide antibiotic, iturin by Bacillus subtilis NB22 in solid state fermentation. J Ferment Bioeng 75:23–27
Phae CG, Shoda M, Kubota H (1990) Suppressive effect of Bacillus subtilis and its products on phytopathogenic microorganisms. J Ferment Bioeng 69:1–7
Sambrook J, Russell DW (2001) Molecular cloning: A laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Sandrin C, Peypoux F, Michel G (1990) Coproduction of surfactin and iturin A, lipopeptides with surfactant and antifungal properties, by Bacillus subtilis. Biotechnol Appl Biochem 12:370–375
Szczech M, Shoda M (2006) The effect of mode of application of Bacillus subtilis RB14-C on its efficacy as a biocontrol agent against Rhizoctonia solani. J Phytopathol 154:370–377
Tokuda Y, Ano T, Shoda M (1993) Survival of Bacillus subtilis NB22, an antifungal–antibiotic iturin A producer, and its transformant in soil-systems. J Ferment Bioeng 75:107–111
Tsuge K, Akiyama T, Shoda M (2001) Cloning, sequencing, and characterization of the iturin A operon. J Bacteriol 183:6265–6273
Yao S, Gao X, Fuchsbauer N, Hillen W, Vater J, Wang J (2003) Cloning, sequencing, and characterization of the genetic region relevant to biosynthesis of the lipopeptides iturin A and surfactin in Bacillus subtilis. Curr Microbiol 47:272–277
Yu GY, Sinclair J B, Hartman GL, Bertagnolli BL (2002) Production of iturin A by Bacillus amyloliquefaciens suppressing Rhizoctonia solani. Soil Biol Biochem 34:955–963
Acknowledgment
This work was supported by the Council of Agriculture grants 92AS-4.2.3-PI-P1 and 93AS-4.1.5-PI-P1.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Hsieh, FC., Lin, TC., Meng, M. et al. Comparing Methods for Identifying Bacillus Strains Capable of Producing the Antifungal Lipopeptide Iturin A. Curr Microbiol 56, 1–5 (2008). https://doi.org/10.1007/s00284-007-9003-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-007-9003-x