Abstract
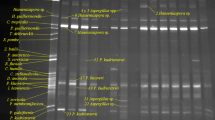
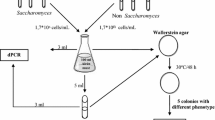
Monitoring for wild yeast contaminants is an essential component of the management of the industrial fuel ethanol manufacturing process. Here we describe the isolation and molecular identification of 24 yeast species present in bioethanol distilleries in northeast Brazil that use sugar cane juice or cane molasses as feeding substrate. Most of the yeast species could be identified readily from their unique amplification-specific polymerase chain reaction (PCR) fingerprint. Yeast of the species Dekkera bruxellensis, Candida tropicalis, Pichia galeiformis, as well as a species of Candida that belongs to the C. intermedia clade, were found to be involved in acute contamination episodes; the remaining 20 species were classified as adventitious. Additional physiologic data confirmed that the presence of these major contaminants cause decreased bioethanol yield. We conclude that PCR fingerprinting can be used in an industrial setting to monitor yeast population dynamics to early identify the presence of the most important contaminant yeasts.

Similar content being viewed by others
References
Abbott DA, Hynes SH, Ingledew WM (2005) Growth rates of Dekkera/Brettanomyces yeasts hinder their ability to compete with Saccharomyces cerevisiae in batch corn mash fermentations. Appl Microbiol Biotechnol 66:641–647
Capece A, Salzano G, Romano P (2003) Molecular typing techniques as a tool to differentiate non-Saccharomyces wine species. Int J Food Microbiol 84:33–39
de Azeredo LA, Gomes EA, Mendonca-Hagler LC, Hagler AN (1998) Yeast communities associated with sugarcane in Campos, Rio de Janeiro, Brazil. Int Microbiol 1:205–208
de Souza-Liberal AT, da Silva Filho EA, de Morais JOF, Simões DA, de Morais Jr MA (2005) Contaminant yeast detection in industrial ethanol fermentation must by rDNA-PCR. Lett Appl Microbiol 40:19–23
de Souza-Liberal AT, Basílio ACM, Brasileiro BTRV, Silva-Filho EA, Simões DA, de Morais Jr MA (2007) Identification of the yeast Dekkera bruxellensis as major contaminant in continuous fuel ethanol fermentation. J App Microbiol 102:538–547
Esteve-Zarzoso B, Belloch C, Uruburu F, Querol A (1999) Identification of yeasts by RFLP analysis of the 5.8S rRNA gene and two ribosomal internal transcribed spacers. Int J Syst Bacteriol 49:329–337
Kurtzman CP, Robnett CJ (1998) Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie van Leeuwenhoek 73:331–371
Loureiro V, Malfeito-Ferreira M (2003) Spoilage yeasts in the wine industry. Int J Food Microbiol 86:23–50
Olasupo NA, Bakre S, Teniola OD, James SA (2003) Identification of yeasts isolated from Nigerian sugar cane peels. J Basic Microbiol 43:530–533
Pina C, Teixeira P, Leite P, Villa M, Belloch C, Brito L (2005) PCR-fingerprinting and RAPD approaches for tracing the source of yeast contamination in a carbonated orange juice production chain. J Appl Microbiol 98:1107–1114
Schwan RF, Mendonca AT, da Silva Jr JJ, Rodrigues V, Wheals AE (2001) Microbiology and physiology of Cachaça (Aguardente) fermentations. Antonie Van Leeuwenhoek 79:89–96
Schuller D, Valero E, Dequin S, Casal M (2004) Survey of molecular methods for the typing of wine yeast strains. FEMS Microbiol Lett 231:19–26
Sherata AM (1960) Yeasts isolated from sugar cane and its juice during the production of aguardente de cana. Appl Microbiol 8:73–75
Silva-Filho EA, Santos SKB, Resende AM, Morais JOF, Morais Jr MA, Simões DA (2005a) Yeast population dynamics of industrial fuel-ethanol fermentation process assessed by PCR-fingerprinting Antonie van Leeuwenhoek 88:13–23
Silva-Filho EA, Melo HF, Antunes DF, dos Santos SKB, Resende MA, Simões DA (2005b) Isolation by genetics and physiological characteristics of a fuel-ethanol fermentative S. cerevisiae strain with potential for genetic manipulation. J Ind Microbiol Biotechnol 32:481–486
Acknowledgments
This work was supported by grants from the Brazilian agencies Fundação de Amparo a Pesquisa do Estado de and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and by financial support from Genetech Bioproductivity, Ltd. (Recife, Brazil).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Basílio, A.C.M., de Araújo, P.R.L., de Morais, J.O.F. et al. Detection and Identification of Wild Yeast Contaminants of the Industrial Fuel Ethanol Fermentation Process. Curr Microbiol 56, 322–326 (2008). https://doi.org/10.1007/s00284-007-9085-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-007-9085-5