Abstract
Deschampsia antarctica Desv. (Poaceae) represents one of the two vascular plants that have colonized the Antarctic continent, which is usually exposed to extreme environmental conditions. In this work, we have characterized the endophytic fungi associated with the leaves of D. antarctica. Endophytic fungi were recovered from 91 individual plants from different points of Admiralty Bay at King George Island, Antarctica. A total of 26 fungal isolates were obtained from 273 leaf fragments. All isolates were identified by analysis of the sequences of the internal transcribed spacer region (ITS) of the rDNA. Alternaria and Phaeosphaeria were the most frequent genera associated with the plant. Other fungal isolates were identified as Entrophospora sp. and several undescribed Ascomycete species. An interesting result was obtained for the isolates UFMGCB 215 and UFMGCB 262, which were related to fungi associated with bryophytes present in boreal ecosystems. Some isolates showed low identity in the ITS sequences to sequences of fungal species deposited in GenBank, suggesting that these fungi could be new species. This work is the first report on fungal endophytes associated with leaves of the Antarctic grass D. antarctica.
Similar content being viewed by others
Introduction
The Antarctic continent is considered to be one of the harshest ecosystems in the world and is the second smallest continent after Australia. This continent is like no other place on earth, and in the last years several studies have been conducted in different scientific areas, including studies with bacteria, protists, fungi, bryophytes, plants, and animals. In general, the flora of Antarctica is restricted to few species of moss, lichens, bryophytes (mosses and liverworts), micro- and macroalgae, and vascular plants. Only two angiosperms are present in Antarctic ecosystems: Deschampsia antarctica Desv. (Poaceae) and Colobanthus quitensis (Kunth) Bartl. (Caryophyllaceae). These angiosperms are highly freezing-tolerant plants (Alberdi et al. 2002) and are found on the Northern and Western parts of the Antarctic Peninsula. Many researchers are interested in D. antarctica due to its ability to live in these extreme conditions (Barrientos-Diaz et al. 2008). Some works have reported on the mechanisms by which these plants resist low temperatures (Gielwanowska and Szczuka 2005). We believe these plants are a target for finding interesting extremophile fungal endophytes.
Endophytic fungi are microfungi that live for all, or at least a part of, their lives inter- or intracellularly and asymptomatically in plant parts without causing apparent symptoms of infection (Petrini 1991) and represent an important portion of fungal diversity. Antarctica is one of the last pristine environments on Earth; as such, it offers unique opportunities to study the diversity and ecology of fungi in extreme environments (Malosso et al. 2006). Molecular analysis of the diversity and phylogenetic relationships of fungi obtained from Antarctic soils show that fungal species are distributed among the Basidiomycota, Ascomycota, and Zygomycota phyla (Malosso et al. 2006). Ruisi et al. (2007) showed an important and complete panorama of different fungal species obtained in Antarctic ecosystems; however, microbial communities, especially fungi, that are present in the Antarctic environment are still poorly characterized. The aim of this study was to recover and identify fungal endophytes living in leaves of D. antarctica collected at different points of Admiralty Bay at King George Island, Antarctica.
Material and methods
Study area
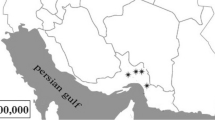
Specimens of Deschampsia antarctica Desv. (Poaceae) were collected in the Antarctic specially managed area (ASMA) in Admiralty Bay, King George Island, South Shetland Islands, Antarctica (62°09′S, 58°28′W). This ASMA was proposed by Brazil and Poland, in coordination with Ecuador and Peru, with the purpose of preventing and/or minimizing the risk of cumulative impacts on the marine and terrestrial environments. Seven separate collection sites were chosen along Admiralty Bay (Fig. 1).
Map of Admiralty Bay at King George Island, Antarctica showing locations (Δ) where samples were obtained for this study (Simões et al. 2004, with modifications)
Plant material and isolation of the endophytic fungi
Fresh leaves of D. antarctica were collected from plants growing under natural conditions in Admiralty Bay during the Antarctic summer in December of 2006 and January of 2007. The leaves were then cut into pieces and stored in plastic bags for no more than 24 h at 10°C before the isolation of endophytic fungi. The leaves were surface sterilized by successively dipping them in ethanol 70% (1 min) and 2% sodium hypochlorite (3 min), followed by washing once with sterile distilled water (2 min) (Collado et al. 1996). Then, the fragments were plated on Petri plates containing potato dextrose agar (PDA, Difco, USA) plus chloramphenicol at 100 μg mL−1. The plates were incubated up to 60 days at 18°C, and individual colonies were transferred to PDA and stored at 4°C. Long-term preservation of mycelial pieces was carried out in cryotubes with 30% sterile glycerol at −80°C. The fungal isolates used in this work were deposited in the Culture Collection of the Departamento of Microbiologia, Instituto of Ciências Biológicas of the Universidade Federal de Minas Gerais (UFMGCB).
The frequency of endophytic fungi growth was calculated by the following formula: N d/N t × 100, where N d and N t are the number of leaves from which the fungi were detected and the total number of leaves, respectively. Percentage abundance of each taxon was calculated according to the following formula: percentage abundance of taxon A = occurrence of taxon A × 100/occurrence of all taxa.
DNA extraction, amplification and sequencing
The protocol for fungal DNA extraction was adapted from De Hoog et al. (2005). The dried DNA was re-suspended in 100 μL TE-buffer (10 mM Tris, 10 mM Na-EDTA, pH 8.0) with 2.5 μL RNAse-solution, which was prepared by adding 10 mg of pancreatic RNAse 20 U mg−1 to 1 mL 0.01 M Na-acetate, heating to 100°C for 15 min, cooling slowly to room temperature, then adjusting the pH to 7.4 with 100 μL Tris–HCl. The sample was incubated for 30 min at 37°C and then stored at −20°C until use (Cota et al. 2008).
The ITS domain of rDNA was amplified with the universal primers ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) as described by White et al. (1990). Amplification of the ITS region was performed as follows: 95°C for 5 min, followed by 40 cycles of 94°C for 60 s, 50°C for 60 s, and 72°C for 60 s, and a final extension at 72°C for 5 min. The amplified DNA was concentrated, cleaned using the Wizard Plus SV Minipreps DNA Purification System (Promega, USA), and sequenced in a MegaBACE™ 1,000 automated sequencing system (Amersham Biosciences, USA). The sequences obtained were adjusted using SeqMan Π of Lasergene software (DNASTAR/Inc.), and a consensus sequence was obtained using the software Bioedit v. 7.0.5.3. To reach species level identification by rDNA sequencing, the consensus sequence was aligned with all sequences of related species retrieved from the GenBank database using the Fasta 2.0 program (Altuschul et al. 1997). After sequencing, the consensus sequences were deposited in the GenBank under the accession numbers EU747133 to EU747158. Phylogenetic relationships were estimated using MEGA program Version 4.0 (Tamura et al. 2007). The phylogenetic tree was constructed using the neighbour joining (NJ) algorithm with bootstrap values calculated from 1,000 iterations. The Kimura 2-parameter model was used to estimate evolutionary distance among the species (Kimura 1980).
Results
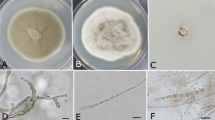
In this study, we used a culture-based approach to examine the endophytic fungi associated with leaves of D. antarctica in different sites of Admiralty Bay. From 91 individual D. antarctica specimens collected, only 26 isolates of fungal endophytes were recovered from 273 leaf fragments, corresponding to an isolation frequency of 9.5%. The 26 endophyte isolates were grouped into five distinct morphospecies taxonomic units. Identification by ITS rDNA sequencing of all endophyte isolates showed that the most abundant fungal taxa belong to the genus Alternaria Nees (57.7%), followed by Phaeosphaeria I. Miyake (19.2%), which were obtained from samples of four and three distinct sites, respectively. The identification of six isolates was not conclusive at taxonomic genus level (Table 1).
A total of 15 isolates were identified as belonging to the genus Alternaria. These isolates showed between 82 and 98% nucleotide sequence identity with Alternaria triticina Prasada & Prabhu (Table 1). These fungi formed a cluster supported by an 84% bootstrap value with Alternaria species but presented 148 (26.2%) and 154 (27.3%) nucleotide differences with A. triticina IA245 and the type strain of A. alternata ATCC 13963, respectively (Fig. 2). The isolates UFMGCB 60, 61, 62, 63, and 102 had identical ITS sequences, and the isolates UFMGCB 103 and 214 differed from them by only one nucleotide (Fig. 2). These isolates were identified as Alternaria sp. 7. The isolates UFMGCB 64, 65, 356, and 360 were very similar with less of 1% difference in the ITS region among them and were identified as Alternaria sp. 5. The isolates UFMGCB 355 and UFMGCB 359, identified as Alternaria sp. 1 and Alternaria sp. 2, respectively, formed a separate group in this cluster. These isolates showed 13.6% nucleotide differences between them. The isolates UFMGCB 261 and UFMGCB 216 presented 4.2% nucleotide differences between them in the sequences of the ITS region and were identified as Alternaria sp. 3 and Alternaria sp. 4, respectively.
The isolates UFMGCB 377 and UFMGCB 378 presented 97% identity in ITS rDNA sequences with Entrophospora sp. (Genbank access number AY035665), a mycorrhizic fungus (Table 1). This isolate, however, presented six nucleotides differences when compared with both Entrophospora sp. and Phaeosphaeria pontifomis. The isolates UFMGCB 213, 215, 357, 358, and 379 were similar in the ITS rDNA sequences (range to 92–98%) with genus Phaeosphaeria, with a 64% bootstrap value, and 9% of differences in the ITS region to P. dennisiana Leuchtm. (Fig. 2). The isolates UFMGCB 213 and 379 showed 2% nucleotide differences in the ITS region between them, and the isolates UFMGCB 215, 357, and 358 presented seven (1.2%) nucleotides among them. The isolate UFMGCB 215 was identified as Phaeosphaeria sp. 2, since it presented 91% sequence identity with Phaeosphaeria sp. B3B (Genbank access number EF432300). This isolate also presented 93% identity in the ITS rDNA region but only 30% of the query coverage with an undescribed fungal endophytic isolate (Genbank access number DQ979616) recovered from moss Huperzia selago (Linnaeus) Bernhardi ex Schrank & Martius. The isolates UFMGCB 380 and UFMGCB 381 presented 97 and 93% identity in the ITS rDNA region sequences, respectively, with an uncultured fungus (Genbank access number EF434086).
The phylogenetic analysis of the isolate UFMGCB 262 showed that it is related to an uncultured fungus (Genbank access number AM999748) recovered from a bryophyte in the boreal forest. This fungal endophyte from D. antarctica showed 16.6% nucleotide differences in the ITS region when compared to the uncultured fungus from the bryophyte.
The isolate UFMGCB 59 was grouped into high taxonomic levels when its sequence was compared to sequences in Genbank. The taxa most related to this isolate presented 85% identity and only 21% of query coverage in the ITS rDNA sequences with an unidentified Ascomycete fungus (Genbank access number DQ683971) obtained from the hyperarid Atacama desert, Chile.
Discussion
Our results raise a number of questions with respect to the diversity of fungi associated with Antarctic plants (angiosperms, moss, and bryophytes). Are there fungal species recorded that are unique to the Antarctic grass studied? How similar is its endophytic fungal community to those grasses living in temperate and tropical regions? Saikkonen et al. (2000), who studied endophytic fungi from wild and cultivated grasses, obtained a low incidence of endophytes from Deschampsia flexuosa and D. cespitosa in extreme drought and cold regions of Finland. Their data are similar to our results and support the hypothesis that the low frequency of endophytic isolates as well as fungal diversity found in D. antarctica leaves may be due to the extreme conditions of Antarctic ecosystems.
According to Petrini (1986), the major group of fungal endophytes is represented by species of Ascomycota and their anamorphs, which were found in this study. The most frequent genus found associated with D. antarctica is Alternaria, a polyphyletic genus classified in the class Hyphomycetes, order Moniliales, and family Dematiaceae (Thomma 2003). This genus is widely distributed in decaying organic materials as well as in many food and feed crops and wild plants and has approximately 50 known species (Kirk et al. 2001). Species of this genus are mainly saprophytes that are commonly found in soil or on decaying plant tissues; some of them have acquired pathogenic capacities (Thomma 2003). Many recent studies have also shown that species belonging to the Alternaria genus are true endophytes associated with temperate and tropical plants (Morakotkarn et al. 2006; Wang et al. 2007), and our study demonstrated their presence in an Antarctic grass. The identification using BLASTn searches (Altuschul et al. 1997) comparing the sequences of the ITS1-5.8S-ITS2 regions of rDNA has shown that the species most related to our Alternaria isolates is A. triticina. This species is known to cause significant yield losses in wheat on the Indian subcontinent, from where it originated and has spread throughout the world (Prasada and Prabhu 1962). The molecular analyses of all Alternaria isolates from D. antarctica showed significant differences, however, from A. triticina and A. alternata. These Antarctic endophytic isolates are probably new species belonging to the genus Alternaria.
The second fungal genus most abundant in leaves of D. antarctica is Phaeosphaeria. This genus is known as a fungal pathogen that causes leaf blotch diseases on wheat. Ernst et al. (2003), however, recovered from a perennial grass species Phragmites australis, an endophytic fungus of the genus Stagonospora, which is a teleomorph stage of Phaeosphaeria. The most related species to our endophyte Phaeosphaeria isolates is P. dennisiana, but the differences in the sequences of the ITS region between the fungal endophytes and the last species suggest that they also represent new species.
The isolate UFMGCB 59 was found to be related to an Ascomycete fungus obtained from the hyperarid Atacama Desert in Chile (Conley et al. 2006). The nucleotide sequence obtained from this isolate, however, was not sufficient to identify it in a specific hierarchical taxonomic level, and more molecular studies are necessary to correctly identify this fungus.
Interesting results were obtained for the isolates UFMGCB 215 and UFMGCB 262, which presented sequence identities to a fungus obtained from a moss living in boreal ecosystems. The isolate UFMGCB 215 showed 93% sequence identity in the ITS region to an undescribed fungal endophytic isolate recovered from Huperzia selago (fir clubmoss; Lycopodiaceae), an herbaceous with evergreen microphylls collected at the boreal forest at Schefferville, Québec, Canada (Higgins et al. 2007), a subartic region. The isolate UFMCB 215 was also found included in the Phaeosphaeria group (Fig. 2), since it presented 91% sequence identity to the endophytic fungus Phaeosphaeria sp. B3B recovered from Brassica nigra (black mustard). The isolate UFMGCB 262 is related to an uncultured fungus (78% identity) obtained from bryophytes of a boreal ecosystem. This isolate also presented low identity to an ectomycorrhizal fungus obtained from the moss Hennediella heimii (Hedw.) Zand. in Dry Valleys of McMurdo Sound, Antarctic. These results show that more work is necessary for correct taxonomic identification of the endophytic fungi associated with D. antarctica.
All fungal endophytes were obtained from leaves of D. antarctica after the Antarctic winter. A total of 21 isolates (80.7%) were black moulds, which are able to produce melanin in their hyphae. The melanin is darkly pigmented polymers that can protect microorganisms against a broad range of environmental stresses; according to Robinson (2001), this compound could protect fungi from extreme temperatures and play a significant role in their persistence from year to year in the environment. Recently, some research has shown that melanised fungi increase their growth after exposure to ionizing radiation, raising questions about a potential role for melanin in energy capture and utilization by black moulds (Dadachova et al. 2007).
According to Ruisi et al. (2007), geographic isolation, combined with environmental stress, make Antarctica a potential place to search for endemic and new fungal species. Studies of endophytic fungi in relatively extreme environments and from phylogenetically distinct lineages of plants are promising sources to recover novel species, which will be important in turn for understanding fungal diversity, the cryptic ecology of microfungal symbionts, and the evolution of plant-fungal symbioses (Higgins et al. 2007). More taxonomic studies are necessary to clarify the correct identification of the fungal endophytes from D. antarctica obtained in our work. Several reports have characterized endophytes fungi from plants living in temperate and tropical regions; however, this study is the first report on endophytic fungi present in leaves from the Antarctic plant D. antarctica.
References
Alberdi M, Bravo LA, Gutiérrez A, Gidekel M, Corcuera LJ (2002) Ecophysiology of Antarctic vascular plants. Physiol Plant 115:479–486
Altuschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Barrientos-Diaz L, Gidekel M, Gutierrez-Moraga A (2008) Characterization of rhizospheric bacteria isolated from Deschampsia antarctica Desv. World J Microbiol Biotechnol 24 (in press)
Collado J, Platas G, Peláez F (1996) Fungal endophytes in leaves, twigs and bark of Quercus ilex from Central Spain. Nova Hedwigia 63:347–360
Conley CA, Ishkhanova G, Mckay CP, Cullungs K (2006) A preliminary survey of non-lichenized fungi cultured from the hyperarid Atacama Desert of Chile. Astrobiology 6:521–526
Cota BB, Rosa LH, Caligiorne RB, Rabello A, Rosa CA, Zani C (2008) Altenusin, a biphenyl isolated from the endophytic fungus Alternaria sp. inhibits trypanothione reductase from Trypanosoma cruzi. FEMS Microbiol Lett 285:177–182
Dadachova E, Bryan RA, Huang X, Ortiz G, Moadel T, Casadevall A (2007) Ionizing radiation changes the electronic properties of melanin and enhances the growth of melanized fungi. PLoS ONE 2:e457
De Hoog GS, Gottlich E, Platas G, Genilloud O, Leotta G, van Brummelen J (2005) Evolution, taxonomy and ecology of the genus Thelebolus in Antarctica. Stud Mycol 51:33–76
Ernst M, Mendgen KW, Wirsel SGR (2003) Endophytic fungal mutualists: seed-borne Stagonospora spp. enhance reed biomass production in axenic microcosms. Mol Plant–Microbe Interactions 16:580–587
Gielwanowska I, Szczuka E (2005) New ultrastructural features of organelles in leaf cells of Deschampsia Antarctica Desv. Polar Biol 28:951–955
Higgins KL, Arnold AE, Miadlikowska J, Sarvate SD, Lutzoni F (2007) Phylogentic relationship, host affinity, and geographic structure of boreal and arctic endophytes from three major plant lineages. Mol Phyl Evol 42:543–555
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kirk PM, Cannon PF, David JC, Stalpers JA (2001) Ainsworth & Bisby’s Dictionary of the Fungi, 19th edn. CAB International, Wallingford, p 655
Malosso E, Waite IS, English L, Hopkins DW, O’Donnel AG (2006) Fungal diversity in maritime Antarctic soils determined using a combination of culture isolation, molecular fingerprinting and cloning techniques. Polar Biol 29:552–561
Morakotkarn D, Kawasaki H, Seki T (2006) Molecular diversity of bamboo-associated fungi isolated from Japan. FEMS Microbiol Lett 266:10–19
Petrini O (1986) Taxonomy of endophytic fungi of aerial plant tissues. In: Fokkema NJ, van den Huevel J (eds) Microbiology of the phyllosphere. Cambridge University Press, England, pp 175–187
Petrini O (1991) Fungal endophytes of tree leaves. In: Andrew JH, Hirano SS (eds) Microbial ecology of leaves. Springer, New York, pp 179–197
Prasada R, Prabhu AS (1962) Leaf blight of wheat caused by a new species of Alternaria. Indian Phytopathol 15:292–293
Robinson CH (2001) Cold adaptation in Artic and Antarctic fungi. New Phytol 151:341–353
Ruisi S, Barreca D, Selbmann L, Zucconi L, Onofri S (2007) Fungi in Antarctica. Rev Environ Sci Biotechnol 6:127–141
Saikkonen K, Ahlholm J, Helander M, Lehtimaki S, Niemelainen O (2000) Endophytic fungi in wild and cultivated grasses in Finland. Ecography 23:360–366
Simões JC, Arigony-Neto J, Bremer UF (2004) O uso de mapas antárticos em publicações. Pesquisa Antartica Brasileira 4:191–197
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Thomma BPHJ (2003) Alternaria spp.: from general saprophyte to specific parasite. Mol. Plant Pathol 4:225–236
Wang B, Priest MJ, Davidson A, Brubaker CL, Woods MJ, Burdon JJ (2007) Fungal endophytes of native Gossypium species in Australia. Mycol Res 111:347–354
White TJ, Bruns TD, Lee SB (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis NA, Gelfand J, Sninsky J et al (eds) PCR protocols: a guide to methods and applications. Academic Press, San Diego, 1989:315–322
Acknowledgments
This study was made possible with financial and logistic support from the Brazilian Antarctic Programme (PROANTAR). It is part of the API activity 403 “MIDIAPI Microbial Diversity of Terrestrial and Maritime ecosystems in Antarctic Peninsula” under the overall coordination of Dr. Vivian H. Pellizari, and contributes to the umbrella IPY activities MERGE (Microbiological and Ecological Responses to Global Environmental Changes in Polar Regions), CAML (Census of Antarctic Marine Life) and SCARMarBIN (SCAR-Marine Biodiversity Information Network).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Rosa, L.H., Vaz, A.B.M., Caligiorne, R.B. et al. Endophytic fungi associated with the Antarctic grass Deschampsia antarctica Desv. (Poaceae). Polar Biol 32, 161–167 (2009). https://doi.org/10.1007/s00300-008-0515-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00300-008-0515-z