Abstract
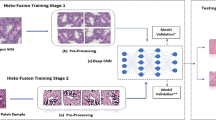
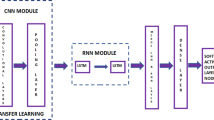
Breast cancer is one of the leading death cause among women nowadays. Several methods have been proposed for the detection of breast cancer. Various machine learning-based automatic diagnosis systems have been developed known as Computer Aided Diagnostics (CAD) systems. Initial CAD systems were based on machine learning algorithms however due to the automatic feature extraction ability of convolutional neural networks (CNN)-based deep learning models are widely adopted. Deep learning is widely used in various fields. Healthcare is one of the essential field that deep learning has transformed. Another common issue faced by the patients is difference of opinion among different pathologists and medical practitioners. Such human errors often lead to misleading or delayed judgment, which may be fatal to human life. To improve decision consistency, efficiency, and error reduction, researchers in the field of healthcare are using deep learning-based approaches and achieved state of art results. In this study, a deep learning (DL) and transfer learning-based approach is proposed to classify histopathological images for breast cancer diagnosis. In this study, we have adopted patch selection approach to classify breast histopathological images on small number of training images using transfer learning without losing the performance. Initially, patches are extracted from Whole Slide Images and fed into the CNN for features extraction. Based on these features, the discriminative patches are selected and then fed to Efficient-Net architecture pre-trained on ImageNet dataset. Features extracted from Efficient-Net architecture are also used to train a SVM classifier. The proposed model outperforms the baseline methods in terms of multiple performance measures.





















Similar content being viewed by others
References
American cancer society.about breast cancer.org—1.800.227.2345. https://www.cancer.org/content/dam/CRC/PDF/Public/8577.00.pdf
Deniz, E., Şengür, A., Kadiroglu, Z., Guo, Y., B̆ajaj, V., Budak, Ü.: Transfer learning based histopathologic image classification for breast cancer detection. Health Inf Sci Syst 6(1), 18 (2018)
Dr.filiz yenicesu.meme kanserinde görüntüleme yöntemleri. https://www.duzen.com.tr/workshop/2011/Meme_Kanserinde_Goruntuleme_Y%C3%B6ntemleri_(Dr_Filiz_Yenicesu).pdf
Das, K., Conjeti, S., Roy, A.G., Chatterjee, J., Sheet, D.: Multiple instance learning of deep convolutional neural networks for breast histopathology whole slide classification. In: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), pp. 578–581. IEEE (2018)
Araújo, T., Aresta, G., Castro, E., Rouco, J., Aguiar, P., Eloy, C., Polónia, A., Campilho, A.: Classification of breast cancer histology images using convolutional neural networks. PLoS ONE 12(6), e0177544 (2017)
Jiang, Y., Chen, L., Zhang, H., Xiao, X.: Breast cancer histopathological image classification using convolutional neural networks with small se-resnet module. PLoS ONE 14(3), e0214587 (2019)
Shen, D., Guorong, W., Suk, H.-I.: Deep learning in medical image analysis. Annu. Rev. Biomed. Eng. 19, 221–248 (2017)
LeCun, Y., Bengio, Y., Hinton, G.: Deep learning. Nature 521(7553), 436–444 (2015)
Anthimopoulos, M., Christodoulidis, S., Ebner, L., Christe, A., Mougiakakou, S.: Lung pattern classification for interstitial lung diseases using a deep convolutional neural network. IEEE Trans Med Imaging 35(5), 1207–1216 (2016)
Shin, H.-C., Roth, H.R., Gao, M., Le, L., Ziyue, X., Nogues, I., Yao, J., Mollura, D., Summers, R.M.: Deep convolutional neural networks for computer-aided detection: Cnn architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging 35(5), 1285–1298 (2016)
Razavian, A.S., Azizpour, H., Sullivan, J., Carlsson, S.: Cnn features off-the-shelf: an astounding baseline for recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition workshops, pp. 806–813 (2014)
Penatti, O.A.B., Nogueira, K., Dos Santos, J.A.: Do deep features generalize from everyday objects to remote sensing and aerial scenes domains? In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, pp. 44–51 (2015)
Tajbakhsh, N., Shin, J.Y., Gurudu, S.R., Todd Hurst, R., Kendall, C.B., Gotway, M.B., Liang, J.: Convolutional neural networks for medical image analysis: full training or fine tuning? IEEE Trans. Med. Imaging 35(5), 1299–1312 (2016)
Mehra, R., et al.: Breast cancer histology images classification: training from scratch or transfer learning? ICT Exp. 4(4), 247–254 (2018)
Akhtar, Z., Foresti, G.L.: Face spoof attack recognition using discriminative image patches. J. Electr. Comput. Eng. 2016, (2016)
Samah, A.A., Fauzi, M.F.A., Mansor, S.: Classification of benign and malignant tumors in histopathology images. In: 2017 IEEE International Conference on Signal and Image Processing Applications (ICSIPA), pp. 102–106. IEEE (2017)
Spanhol, F.A., Oliveira, L.S., Petitjean, C., Heutte, L.: A dataset for breast cancer histopathological image classification. IEEE Trans. Biomed. Eng. 63(7), 1455–1462 (2015)
Kahya, M.A., Al-Hayani, W., Algamal, Z.Y.: Classification of breast cancer histopathology images based on adaptive sparse support vector machine. J. Appl. Math. Bioinform. 7(1), 49 (2017)
Sanchez-Morillo, D., González, J., García-Rojo, M., Ortega, J.: Classification of breast cancer histopathological images using kaze features. In: International Conference on Bioinformatics and Biomedical Engineering, pp. 276–286. Springer (2018)
Alcantarilla, P.F., Bartoli, A., Davison, A.J.: Kaze features. In: European Conference on Computer Vision, pp. 214–227. Springer (2012)
Chan, A., Tuszynski, J.A.: Automatic prediction of tumour malignancy in breast cancer with fractal dimension. R. Soc. Open Sci. 3(12), 160558 (2016)
Nejad, E.M., Affendey, L.S., Latip, R.B., Ishak, I.B.: Classification of histopathology images of breast into benign and malignant using a single-layer convolutional neural network. In: Proceedings of the International Conference on Imaging, Signal Processing and Communication, pp. 50–53 (2017)
Nahid, A.-A., Mehrabi, M.A., Kong, Y.: Histopathological breast cancer image classification by deep neural network techniques guided by local clustering. BioMed Research International (2018)
Kumar, K., Rao, A.C.S.: Breast cancer classification of image using convolutional neural network. In: 2018 4th International Conference on Recent Advances in Information Technology (RAIT), pp. 1–6. IEEE (2018)
Sun, J., Binder, A.: Comparison of deep learning architectures for h&e histopathology images. In: 2017 IEEE Conference on Big Data and Analytics (ICBDA), pp. 43–48. IEEE (2017)
Benhammou, Y., Tabik, S., Achchab, B., Herrera, F.: A first study exploring the performance of the state-of-the art cnn model in the problem of breast cancer. In: Proceedings of the International Conference on Learning and Optimization Algorithms: Theory and Applications, pp. 1–6 (2018)
Motlagh, N.H., Jannesary, M., Aboulkheyr, H., Khosravi, P., Elemento, O., Totonchi, M., Hajirasouliha, I.: Breast cancer histopathological image classification: a deep learning approach. bioRxiv, p. 242818 (2018)
Nawaz, M.A., Sewissy, A.A., Soliman, T.H.A.: Automated classification of breast cancer histology images using deep learning based convolutional neural networks. Int. J. Comput. Sci. Netw. Secur. 4, 152–160 (2018)
Sharma, S., Mehra, R.: Conventional machine learning and deep learning approach for multi-classification of breast cancer histopathology images-a comparative insight. J. Digit. Imaging 33(3), 632–54 (2020)
Guo, Y., et al.: DeepANF: a deep attentive neural framework with distributed representation for chromatin accessibility prediction. Neurocomputing 379, 305–318 (2020)
Lyu, C., Wang, L., Zhang, J.: Deep learning for dnase i hypersensitive sites identification. BMC Genom. 19(10), 905 (2018)
Guo, Y., et al.: Attentive gated neural networks for identifying chromatin accessibility. Neural Comput. Appl. (2020)
de Matos, J., et al.: Double transfer learning for breast cancer histopathologic image classification. In: 2019 International Joint Conference on Neural Networks (IJCNN). IEEE (2019)
Singh, J., et al.: RNA secondary structure prediction using an ensemble of two-dimensional deep neural networks and transfer learning. Nat. Commun. 10(1), 1–13 (2019)
Guo, Y., et al.: DeepACLSTM: deep asymmetric convolutional long short-term memory neural models for protein secondary structure prediction. BMC Bioinform. 20(1), 1–12 (2019)
Orenstein, E.C., Beijbom, O.: Transfer learning and deep feature extraction for planktonic image data sets. In: 2017 IEEE Winter Conference on Applications of Computer Vision (WACV), pp. 1082–1088. IEEE (2017)
BreakHis Dataset link. https://web.inf.ufpr.br/vri/databases/breast-cancer-histopathological-database-breakhis/
Wang, Z.F., Xie, Z.F., Qiu, P.C.: Comparison of data standardization method in semantic relation similarity calculation. Comput Eng 38(10), 38–40 (2012)
Tan, M., Le, Q.V.: Efficientnet: rethinking model scaling for convolutional neural networks (2019). arXiv preprint arXiv:1905.11946
Uludag, U., Ross, A., Jain, A.: Biometric template selection and update: a case study in fingerprints. Pattern Recognit. 37(7), 1533–1542 (2004)
Perez, L., Wang, J.: The effectiveness of data augmentation in image classification using deep learning (2017). arXiv preprint arXiv:1712.04621
Kingma, D.P., Ba, J.: Adam: A Method for Stochastic Optimization (2014). arXiv preprint arXiv:1412.6980
Benhammou, Y., Achchab, B., Herrera, F., Tabik, S.: Breakhis based breast cancer automatic diagnosis using deep learning: taxonomy, survey and insights. Neurocomputing 375, 9–24 (2020)
Han, Z., Wei, B., Zheng, Y., Yin, Y., Li, K., Li, Sh: Breast cancer multi-classification from histopathological images with structured deep learning model. Sci. Rep. 7(1), 1–10 (2017)
Bardou, D., Zhang, K., Ahmad, S.M.: Classification of breast cancer based on histology images using convolutional neural networks. IEEE Access 6, 24680–24693 (2018)
Wang, P., Wang, J., Li, Y., Li, P., Li, L., Jiang, M.: Automatic classification of breast cancer histopathological images based on deep feature fusion and enhanced routing. Biomed. Signal Process. Control 1(65), 102341 (2021)
Nahid, A.-A., Kong, Y.: Histopathological breast-image classification using local and frequency domains by convolutional neural network. Information 9(1), 19 (2018)
Gandomkar, Z., Brennan, P.C., Mello-Thoms, C.: MuDeRN: multi-category classification of breast histopathological image using deep residual networks. Artif. Intell. Med. 88, 14–24 (2018)
Lin, M., Chen, Q., Yan, S.: Network in network (2013). arXiv preprint arXiv:1312.4400
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ahmad, N., Asghar, S. & Gillani, S.A. Transfer learning-assisted multi-resolution breast cancer histopathological images classification. Vis Comput 38, 2751–2770 (2022). https://doi.org/10.1007/s00371-021-02153-y
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00371-021-02153-y