Abstract
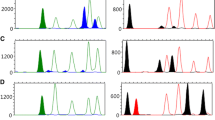
Identification of body fluids found at crime scenes provides important information that can support a link between sample donors and actual criminal acts. Previous studies have reported that DNA methylation analysis at several tissue-specific differentially methylated regions (tDMRs) enables successful identification of semen, and the detection of certain bacterial DNA can allow for identification of saliva and vaginal fluid. In the present study, a method for detecting bacterial DNA was integrated into a previously reported multiplex methylation-sensitive restriction enzyme-polymerase chain reaction. The developed multiplex PCR was modified by the addition of a new semen-specific marker and by including amplicons for the 16S ribosomal RNA gene of saliva- and vaginal fluid-specific bacteria to improve the efficacy to detect a specific type of body fluid. Using the developed multiplex system, semen was distinguishable by unmethylation at the USP49, DACT1, and PFN3 tDMRs and by hypermethylation at L81528, and saliva could be identified by detection of saliva-specific bacteria, Veillonella atypica and/or Streptococcus salivarius. Additionally, vaginal fluid and menstrual blood were differentiated from other body fluids by hypomethylation at the PFN3 tDMR and the presence of vaginal fluid-specific bacteria, Lactobacillus crispatus and/or Lactobacillus gasseri. Because the developed multiplex system uses the same biological source of DNA for individual identification profiling and simultaneously analyses various types of body fluid in one PCR reaction, this method will facilitate more efficient body fluid identification in forensic casework.



Similar content being viewed by others
References
Virkler K, Lednev IK (2009) Analysis of body fluids for forensic purposes: from laboratory testing to non-destructive rapid confirmatory identification at a crime scene. Forensic Sci Int 188:1–17
An JH, Shin KJ, Yang WI, Lee HY (2012) Body fluid identification in forensics. BMB Rep 45:545–553
Zubakov D, Hanekamp E, Kokshoorn M, van IJcken W, Kayser M (2008) Stable RNA markers for identification of blood and saliva stains revealed from whole genome expression analysis of time-wise degraded samples. Int J Legal Med 122:135–142
Hanson EK, Lubenow H, Ballantyne J (2009) Identification of forensically relevant body fluids using a panel of differentially expressed microRNAs. Anal Biochem 387:303–314
Fleming RI, Harbison S (2010) The development of a mRNA multiplex RT-PCR assay for the definitive identification of body fluids. Forensic Sci Int Genet 4:244–256
Frumkin D, Wasserstrom A, Budowle B, Davidson A (2011) DNA methylation-based forensic tissue identification. Forensic Sci Int Genet 5:517–524
Lee HY, Park MJ, Choi A, An JH, Yang WI, Shin KJ (2012) Potential forensic application of DNA methylation profiling to body fluid identification. Int J Legal Med 126:55–62
An JH, Choi A, Shin KJ, Yang WI, Lee HY (2013) DNA methylation-specific multiplex assays for body fluid identification. Int J Legal Med 127:35–43
Wasserstrom A, Frumkin D, Davidson A, Shpitzen M, Herman Y, Gafny R (2013) Demonstration of DSI-semen—a novel DNA methylation-based forensic semen identification assay. Forensic Sci Int Genet 7:136–142
Larue BL, King JL, Budowle B (2013) A validation study of the Nucleix DSI-Semen kit—a methylation-based assay for semen identification. Int J Legal Med 127:299–308
Madi T, Balamurugan K, Bombardi R, Duncan G, McCord B (2012) The determination of tissue-specific DNA methylation patterns in forensic biofluids using bisulfite modification and pyrosequencing. Electrophoresis 33:1736–1745
Nakanishi H, Kido A, Ohmori T, Takada A, Hara M, Adachi N et al (2009) A novel method for the identification of saliva by detecting oral streptococci using PCR. Forensic Sci Int 183:20–23
Nakanishi H, Ohmori T, Hara M, Takada A, Shojo H, Adachi N et al (2011) A simple identification method of saliva by detecting Streptococcus salivarius using loop-mediated isothermal amplification. J Forensic Sci 56(Suppl 1):S158–S161
Hsu L, Power D, Upritchard J, Burton J, Friedlander R, Horswell J et al (2012) Amplification of oral streptococcal DNA from human incisors and bite marks. Curr Microbiol 65:207–211
Akutsu T, Motani H, Watanabe K, Iwase H, Sakurada K (2012) Detection of bacterial 16S ribosomal RNA genes for forensic identification of vaginal fluid. Legal Med (Tokyo) 14:160–162
Benschop CC, Quaak FC, Boon ME, Sijen T, Kuiper I (2012) Vaginal microbial flora analysis by next generation sequencing and microarrays; can microbes indicate vaginal origin in a forensic context? Int J Legal Med 126:303–310
Giampaoli S, Berti A, Valeriani F, Gianfranceschi G, Piccolella A, Buggiotti L et al (2012) Molecular identification of vaginal fluid by microbial signature. Forensic Sci Int Genet 6:559–564
Ohgane J, Yagi S, Shiota K (2008) Epigenetics: the DNA methylation profile of tissue-dependent and differentially methylated regions in cells. Placenta 29:S29–S35
Song F, Smith JF, Kimura MT, Morrow AD, Matsuyama T, Nagase H et al (2005) Association of tissue-specific differentially methylated regions (TDMs) with differential gene expression. Proc Natl Acad Sci U S A 102:3336–3341
Kitamura E, Igarashi J, Morohashi A, Hida N, Oinuma T, Nemoto N et al (2007) Analysis of tissue-specific differentially methylated regions (TDMs) in humans. Genomics 89:326–337
Illingworth R, Kerr A, Desousa D, Jørgensen H, Ellis P, Stalker J et al (2008) A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol 6:e22
Chakravorty S, Helb D, Burday M, Connell N, Alland D (2007) A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J Microbiol Methods 69:330–339
De Backer E, Verhelst R, Verstraelen H, Alqumber MA, Burton JP, Tagg JR et al (2007) Quantitative determination by real-time PCR of four vaginal Lactobacillus species, Gardnerella vaginalis and Atopobium vaginae indicates an inverse relationship between L. gasseri and L. iners. BMC Microbiol 7:115
Kang JG, Kim SH, Ahn TY (2006) Bacterial diversity in the human saliva from different ages. J Microbiol 44:572–576
Marinus MG, Morris NR (1973) Isolation of deoxyribonucleic acid methylase mutants of Escherichia coli K-12. J Bacteriol 114:1143–1150
May MS, Hattman S (1975) Analysis of bacteriophage deoxyribonucleic acid sequences methylated by host- and R-factor-controlled enzymes. J Bacteriol 123:768–770
Gajer P, Brotman RM, Bai G, Sakamoto J, Schütte UM, Zhong X et al (2012) Temporal dynamics of the human vaginal microbiota. Sci Transl Med 4:132ra52
Witkin SS, Linhares IM, Giraldo P (2007) Bacterial flora of the female genital tract: function and immune regulation. Best Pract Res Clin Obstet Gynaecol 21:347–354
Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL et al (2011) Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A 108:4680–4687
Fleming RI, Harbison S (2010) The use of bacteria for the identification of vaginal secretions. Forensic Sci Int Genet 4:311–315
Acknowledgments
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology (2010-0005208).
Conflicts of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Choi, A., Shin, KJ., Yang, W.I. et al. Body fluid identification by integrated analysis of DNA methylation and body fluid-specific microbial DNA. Int J Legal Med 128, 33–41 (2014). https://doi.org/10.1007/s00414-013-0918-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00414-013-0918-4