Abstract
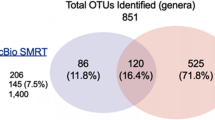
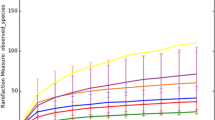
In contrast to conventional wastewater treatment plants and saline environments, little is known regarding the microbial diversity of hypersaline wastewater. In this study, the microbial communities of a hypersaline tannery effluent, and those of three treatment systems operating with the tannery effluent, were investigated using 16S rDNA phylogenetic markers. The comparative analysis of 377 bacterial sequences revealed the high diversity of this type of hypersaline environment, clustering within 193 phylotypes (≥ 97% similarity) and covering 14 of the 52 divisions of the bacterial domain, i.e. Proteobacteria, Bacteroidetes, Firmicutes, Actinobacteria, Chlorobi, Planctomycetes, Spirochaetes, Synergistes, Chloroflexi, Thermotogae, Verrucomicrobia, OP3, OP11 and TM7. Most of the phylotypes were related to halophilic and pollutant-degrading bacteria. Using statistical analysis, the diversity of this type of environment was compared to that of other environmental samples selected on the basis of their salinity, oxygen content and organic load.



Similar content being viewed by others
References
Altschul S, Madden T, Schaffer A, Zhang J, Zhang Z, Miller W, Lipman D (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Anton J, Llobet-Brossa E, Rodriguez-Valera F, Amann R (1999) Fluorescence in situ hybridization analysis of the prokaryotic community inhabiting crystallizer ponds. Environ Microbiol 1:517–523
Anton J, Rossello-Mora R, Rodriguez-Valera F, Amann R (2000) Extremely halophilic Bacteria in crystallizer ponds from solar salterns. Appl Environ Microbiol 66:3052–3057
Benlloch S, Martinez-Murcia AJ, Rodriguez-Valera F (1995) Sequencing of bacterial and archaeal 16S rRNA genes directly amplified from a hypersaline environment. Syst Appl Microbiol 18:574–581
Benlloch S, Lopez-Lopez A, Casamayor EO, Ovreas L, Goddard V, Daae FL, Smerdon G, Massana R, Joint I, Thingstad F, Pedros-Alio C, Rodriguez-Valera F (2002) Prokaryotic genetic diversity throughout the salinity gradient of a coastal solar saltern. Environ Microbiol 4:349–360
Bond PL, Hugenholtz P, Keller J, Blackall LL (1995) Bacterial community structures of phosphate-removing and non-phosphate-removing activated sludges from sequencing batch reactors. Appl Environ Microbiol 61:1910–1916
Brosius J, Dull TJ, Sleeter DD, Noller HF (1981) Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol 148:107–127
Chao A, Hwang WH, Chen YC, Kuo CY (2000) Estimating the number of shared species in two communities. Stat Sin 10:227–246
Chazdon RL, Colwell RK, Denslow JS, Guariguata MR (1998) Statistical methods for estimating species richness of woody regeneration in primary and secondary rain forests of NE Costa Rica. In: Dallmeier F, Comiskey JA (eds) Forest biodiversity research, monitoring and modeling: conceptual background and Old World case studies. Parthenon Publishing, Paris, pp 285–309
Cole JR, Chai B, Marsh TL, Farris RJ, Wang Q, Kulam SA, Chandra S, McGarrell DM, Schmidt TM, Garrity GM et al (2003) The ribosomal database project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res 31:442–443
Colwell RK (2005) EstimateS: statistical estimation of species richness and shared species from samples. Version 7.5. User’s Guide and application published at: http://www.purl.oclc.org/estimates
Duckworth AW, Grant WD, Jones BE, van Steenbergen R (1996) Phylogenetic diversity of soda lake alkaliphiles. FEMS Microbiol Ecol 19:181–191
Edgcomb VP, Kysela DT, Teske A, de Vera Gomez A, Sogin ML (2002) Benthic eukaryotic diversity in the Guaymas Basin hydrothermal vent environment. Proc Natl Acad Sci USA 99:7658–7662
Godon JJ, Zumstein E, Dabert P, Habouzit F, Moletta R (1997a) Microbial 16S rDNA diversity in an anaerobic digester. Water Sci Technol 36:49–55
Godon JJ, Zumstein E, Dabert P, Habouzit F, Moletta R (1997b) Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequence analysis. Appl Environ Microbiol 63:2802–22813
Godon JJ, Bayle S, Leclerc M, Delgenes JP (2004) Comparison of 16S ribosomal DNA structure of ten anaerobic digestors. In: Final programm abstracts, 10th Int Symp Microb Ecol. Cancun, Mexico, p 176
Good IL (1953) The population frequencies of species and the estimation of population parameters. Biometrika 40:237–264
Hollibaugh JT, Wong PS, Bano N, Pak SK, Prager EM, Orrego C (2001) Stratification of microbial assemblages in Mono Lake, California, and response to a mixing event. Hydrobiologia 466:45–60
Humayoun SB, Bano N, Hollibaugh JT (2003) Depth distribution of microbial diversity in Mono Lake, a meromictic soda lake in California. Appl Environ Microbiol 69:1030–1042
Jones BE (2004) Industrial enzymes: do halophiles and alkaliphiles have a role to play? In: Ventosa A (ed) Halophilic microorganisms. Springer, Berlin Heidelberg New York, pp 275–284
Lefebvre O, Habouzit F, Bru V, Delgenes JP, Godon JJ, Moletta R (2004) Treatment of hypersaline industrial wastewater by a microbial consortium in a sequencing batch reactor. Environ Technol 25:543–553
Lefebvre O, Vasudevan N, Torrijos M, Thanasekaran K, Moletta R (2005) Halophilic biological treatment of tannery soak liquor in a sequencing batch reactor. Water Res 39:1471–1480
Lefebvre O, Vasudevan N, Torrijos M, Thanasekaran K, Moletta R (2006) Anaerobic digestion of tannery soak liquor with an aerobic post-treatment. Water Res 40:1492–1500
Litchfield CD (2004) Microbial molecular and physiological diversity in hypersaline environments. In: Ventosa A (ed) Halophilic microorganisms. Springer, Berlin Heidelberg New York, pp 49–61
Litchfield CD, Gillevet PM (2002) Microbial diversity and complexity in hypersaline environments: a preliminary assessment. J Ind Microbiol Biotechnol 28:48–55
Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, Buchner A, Lai T, Steppi S, Jobb G, Forster W et al (2004) ARB: a software environment for sequence data. Nucleic Acids Res 32:1363–1371
Martinez-Murcia AJ, Acinas SG, Rodriguez-Valera F (1995) Evaluation of prokaryotic diversity by restrictase digestion of 16S rDNA directly amplified from hypersaline environments. FEMS Microbiol Ecol 17:247–255
Mouné S, Caumette P, Matheron R, Willison JC (2003) Molecular sequence analysis of prokaryotic diversity in the anoxic sediments underlying cyanobacterial mats of two hypersaline ponds in Mediterranean salterns. FEMS Microbiol Ecol 44:117–130
Peyton BM, Mormile MR, Alva V, Oie C, Roberto F, Apel WA, Oren A (2004) Biotransformation of toxic organic and inorganic contaminants by halophilic bacteria. In: Ventosa A (ed) Halophilic microorganisms. Springer, Berlin Heidelberg New York, pp 315–331
Quesada E, Bejar V, Ferrer MR, Calvo C, Llamas I, Martinez-Checa F, Arias S, Ruiz-Garcia C, Paez R, Martinez-Canovas MJ, Del Moral A (2004) Moderately halophilic, exopolysaccharide-producing bacteria. In: Ventosa A (ed) Halophilic microorganisms. Springer, Berlin Heidelberg New York, pp 297–314
Rappé MS, Giovannoni SJ (2003) The uncultured microbial majority. Annu Rev Microbiol 57:369–394
Rodríguez-Valera F, Acinas SG, Anton J (1999) Contribution of molecular techniques to the study of microbial diversity in hypersaline environments. In: Oren A (ed) Microbiology and biogeochemistry of hypersaline environments. CRC Press, Boca Raton, pp 27–38
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sekiguchi Y, Kamagata Y, Syutsubo K, Ohashi A, Harada H, Nakamura K (1998) Phylogenetic diversity of mesophilic and thermophilic granular sludges determined by 16S rRNA gene analysis. Microbiology 144:2655–2665
Snaidr J, Amann R, Huber I, Ludwig W, Schleifer KH (1997) Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl Environ Microbiol 63:2884–2896
Snell-Castro R, Godon J-J, Delgenes J-P, Dabert P (2005) Characterisation of the microbial diversity in a pig manure storage pit using small subunit rDNA sequence analysis. FEMS Microbiol Ecol 52:229–242
Sorokin DY, Kuenen JG (2005) Haloalkaliphilic sulfur-oxidizing bacteria in soda lakes. FEMS Microbiol Rev 29:685–702
Sreeram KJ, Ramasami T (2003) Sustaining tanning process through conservation, recovery and better utilization of chromium. Resour Conserv Recycling 38:185–212
Stoop MLM (2003) Water management of production systems optimised by environmentally oriented integral chain management: case study of leather manufacturing in developing countries. Technovation 23:265–278
Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu DY, Paulsen I, Nelson KE, Nelson W, Fouts DE, Levy S, Knap AH, Lomas MW, Nealson K, White O, Peterson J, Hoffman J, Parsons R, Baden-Tillson H, Pfannkoch C, Rogers YH, Smith HO (2004) Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66–74
Ward BB, Martino DP, Diaz MC, Joye SB (2000) Analysis of ammonia-oxidizing bacteria from hypersaline Mono Lake, California, on the basis of 16S rRNA sequences. Appl Environ Microbiol 66:2873–2881
Wiegant WM, Kalker TJJ, Sontakke VN, Zwaag RR (1999) Full scale experience with tannery water management: an integrated approach. Water Sci Technol 39:169–176
Acknowledgements
This work was supported by the Indo-French Cell for Bioprocesses on Environment (IFCBE) based on cooperation between the Centre for Environmental Studies, Anna University, Chennai (India) and the French National Institute for Agricultural Research (Institut National de la Recherche Agronomique, INRA). The authors would like to thank Dr. P. Dabert and Ms. V. Bru for their assistance during the study and Dr. M. Moletta for providing data regarding anaerobic sludge diversity.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by K. Horikoshi
Rights and permissions
About this article
Cite this article
Lefebvre, O., Vasudevan, N., Thanasekaran, K. et al. Microbial diversity in hypersaline wastewater: the example of tanneries. Extremophiles 10, 505–513 (2006). https://doi.org/10.1007/s00792-006-0524-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-006-0524-1