Abstract
A simple stochastic approach, designed to model the movement of electrons throughout chemical bonds, is introduced. This model makes use of a Markov matrix to codify useful structural information in QSAR. The self-return probabilities of this matrix throughout time (SRπ k ) are then used as molecular descriptors. Firstly, a calculation of SRπ k is made for a large series of anticancer and non-anticancer chemicals. Then, k-Means Cluster Analysis allows us to split the data series into clusters and ensure a representative design of training and predicting series. Next, we develop a classification function through Linear Discriminant Analysis (LDA). This QSAR discriminates between anticancer compounds and non-active compounds with a correct global classification of 90.5% in the training series. The model also correctly classified 86.07% of the compounds in the predicting series. This classification function is then used to perform a virtual screening of a combinatorial library of coumarins. In this connection, the biological assay of some furocoumarins, selected by virtual screening using the present model, gives good results. In particular, a tetracyclic derivative of 5-methoxypsoralen (5-MOP) has an IC50 against HL-60 tumoral line around 6 to 10 times lower than those for 8-MOP and 5-MOP (reference drugs), respectively. Finally, application of Iso-contribution Zone Analysis (IZA) provides structural interpretation of the biological activity predicted with this QSAR.
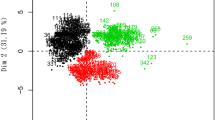
Figure IZA of compound 1







Similar content being viewed by others
References
Markov AA (1906) Bull Soc Phys Math Kasan 15:155–156
Bharucha-Reid AT (1960) Elements of theory of markov process on the application. McGraw-Hill Series in Probability and Statistics. McGraw-Hill, New York, pp 167–434
Freund JA, Poschel T (eds) (2000) Stochastic processes in physics, chemistry, and biology. In: Lecture Notes in Physics. Springer, Berlin Heidelberg New York
Graepel T, Obermayer K (1998) Neural Comput 11:39–55
Geng J, Xu D, Gong J, Li W (1998) Int J Epidemiol 27:320–322
Yakovlev A, Boucher K, DiSario J (1999) Math Biosci 1:45–60
Vorodovsky M, Koonin EV, Rudd KE (1994) Trends Biochem Sci 19:309–313
Vorodovsky M, MacIninch JD, Koonin EV, Rudd KE, Médigue C, Danchin A (1995) Nucleic Acid Res 23:3554–3562
Chou KC (1997) Biopolymers 42:837–853
Yuan Z (1999) FEBS Lett 451:23–26
Hua S, Sun Z (2001) Bioinformatics 17:721–728
Hubbard TJ, Park J (1995) Proteins Struct Funct Genet 23:398–402
Krogh A, Brown M, Mian IS, Sjeander K, Haussler D (1994) J Mol Biol 235:1501–1531
Di Francesco V, Munson PJ, Garnier J (1999) Bioinformatics 15:131–140
James AJ (1995) Solving the many electron problem with quantum Monte-Carlo methods. Imperial College of Science, Technology and Medicine, London, pp 12–202
Landau LD, Lifshitz EM (1963) Mecánica Quántica no-Relativista. In: Curso de Física Teórica, vol 3. Reverté, Barcelona, pp 1–49
Dreizler RM, Gross EKU (1990) Density functional theory: an approach to the quantum many-body problem. Springer, Berlin Heidelberg New York, pp 1–30
Estrada E, Uriarte E (2001) Curr Med Chem 8:1573–1588
Kubinyi H (1999) J Recept Signal Transduct Res 19:15–39
Kier LB, Hall LH (1999) Topological indices and related descriptors in QSAR and QSPR. Gordon and Breach, Amsterdam, pp 455–489
Devillers J, Balaban AT (2000) Topological indices and related descriptors in QSAR and drug design. Amsterdam, pp 3–41
Hall LH, Kier LB (1977) Tetrahedron 33:1953–1957
Bonchev D (1983) Information theoretic indices for characterization of chemical structure. RSP-Wiley, Chichester, UK, pp 4–20
Trinjastic N (1992) Chemical graph theory. CRC Press, Boca Raton, Fla., pp 1–30
Bonchev D, Rouvray DH (1991) Chemical graph theory. Gordon and Breach, New York, pp 6–23
Dewar MJ (1991) MOTEC 91, modern technique in computational chemistry. Leiden, pp 6–15
Ögnetir C, Csizmadia IG (1991) Computational advances in organic chemistry: molecular structure and reactivity. Kluwer, Dordrecht
Kikuchi O (1987) Quant Struct–Act Relat 6:179–184
Gajewski JJ, Gilbert KE, Mckelvey J (1990) Advances in molecular modeling. JAI Press, Greenwich, pp 65–68
Estrada E (1997) J Chem Inf Comput Sci 37:320–328
Estrada E (1996) J Chem Inf Comput Sci 36:844–849
Stewart JJ (1989) J Comput Chem 10:209–221
Dewar MJ, Zoebish EG, Healy EF, Stewart JJ (1985) J Am Chem Soc 107:3902–3909
Todeschini R, Consonni V (2000) Handbook of molecular descriptors. Wiley-VCH, Weinheim
Denny WA (1992) The role of medicinal chemistry in the discovery of DNA-active anticancer drugs: from random searching, through lead development, to the novo design. In: Waring MJ, Ponder BAJ (eds) The search for anticancer drugs. Kluwer, Dordrecht, chapter 2
Lunney EA (1998) Med Chem Res 8:352–361
Walters WP, Stahl MT, Murcko MA (1998) Drug Discovery Today 3:160–178
Kubinyi H (1999) J Recept Signal Transduct Res 19:15–39
Lien EJ, Lien LL (1998) Chin Phar J 50:249–256
Lunney EA (1998) Med Chem Res 8:352–361
Walters WP, Stahl MT, Murcko MA (1998) Drug Discovery Today 3:160–178
Drie JHV, Lajines MS (1998) Drug Discovery Today 3:274–283
Ferrante K, Winograd B, Canetta R (1999) Cancer Chemother Pharmacol 43:S61-S83
Menta E, Palumbo M (1998) Expert Opin Ther Pat 8:1627–1672
Estrada E, Gutiérrez Y, González DH (2000) J Chem Inf Comput Sci 40:1386–1399
Estrada E, Gonzalez DH (2003) J Chem Inf Comput Sci 43:75–84
Randič M (1991) J Math Chem 7:155–168
Padron JA, Carrasco R, Pellón RF (2002) J Pharm Pharmaceut Sci 5:267–274
González DH, Olazábal E, Castañedo N, Hernádez SI, Morales A, Serrano HS, González J, Ramos de Armas R (2002) J Mol Mod 8:237–245
González DH, Hernádez SI, Uriarte E, Santana L (2003) Comput Chem (in press, corrected proofs published online)
González DH, De Armas RR, Uriarte E (2002) Online J Bioinformatics 1:83–95
Cabrera MA, González DH, Teruel C, Pla-Delfina JM, Bermejo del Val M (2002) Eur J Pharm Biopharm 53:317–325
Randič M (1991) Chemom Intell Lab Sist 10:213–227
Hall LH, Mohney B, Kier LB (1991) Quant Struct–Act Relat 10:43–51
Gálvez J, García R, Salabert MT, Soler R (1994) J Chem Inf Comput Sci 34:520–525
Gnedenko B (1978) The theory of probability. Mir, Moscow, pp 107–112
Pauling L (1939) The nature of the chemical bond. Cornell University Press, Ithaca, N.Y., pp 2–60
Grimmett GR, Stirzaker DR (1992) Probability and random processes. Clarendon Press, Oxford, pp 194–264
Kier LB, Hall LH (1999) Molecular structure description. The electrotopological state. Academic Press, New York
Estrada E, Molina E (2001) J Chem Inf Comput Sci 41:791-797
Hernández I, González H (2002) MARCH-INSIDE version 1.0 (Markovian chemicals "in silico" design). Chemicals Bio-actives Center, Central University of Las Villas, Cuba. This is a preliminary experimental version future professional version shall be available to the public. For any information about it, send an e-mail to the corresponding author humbertogd@cbq.uclv.edu.cu
Rogers KS, Camnarata A (1969) J Med Chem 12:692–693
Jiang Y, Tang A, Hoffman R (1984) Theor Chim Acta 66:183–192
Burdett JK, Lee S (1985) J Am Chem Soc 107:3063–3082
Burdett JK, Lee S (1985) J Am Chem Soc 107:3050–3063
Lee S (1991) Acc Chem Res 24:249–254
Markovick S, Gutman I (1991) J Mol Struct (THEOCHEM) 81:81–87
Gutman I, Rosenfield VR (1996) Theor Chim Acta 93:191–197
Gutman I (1992) Theor Chim Acta 83:313–318
Karwowski J, Bielinska-Waz D, Jurkowski J (1996) Int Quantum Chem 60:185–193
Estrada E, Peña A, García-Domenech R (1998) J Comp Aided Mol Design 12:583–595
STATISTICA for Windows (2001) release 6.0. Statsoft Inc
van Waterbeemd H (1995) Discriminant analysis for activity prediction. In: Manhnhold R, Krogsgaard-Larsen P, Timmerman H (eds) Method and principles in medicinal chemistry, vol 2. Chemometric methods in molecular design, van Waterbeemd H (ed). VCH, Weinhiem, pp 265–282
Julián-Ortiz JV, Gálvez J, Mullños-Collado C, García-Domenech R, Gimeno-Cardona C (1999) J Med Chem 42:3308–3314
Kowalski RB, Wold S (1982) Pattern recognition in chemistry. In: Krishnaiah PR, Kanal LN (eds) Handbook of statistics. North-Holland, Amsterdam, pp 673–697
Mc Farland JW, Cooper CB, Newcomb DM (1991) J Med Chem 34:1908–1911
Negwer M (1987) Organic chemical drugs and their synonyms. Akademie-Verlag, Berlin
Mc Farland JW, Gans DJ (1995) Cluster significance analysis. In: Manhnhold R, Krogsgaard-Larsen P, Timmerman H (eds) Method and principles in medicinal chemistry, vol 2. Chemometric methods in molecular design, van Waterbeemd H (ed). VCH, Weinhiem, pp 295–307
Johnson RA, Wichern DW (1988) Applied multivariate statistical analysis. Prentice-Hall, N.J.
Via DL, Gia O, Magno MS, Da Settimo A, Primofiore G, Da Settimo F, Simorini F, Marini AM (2002) Eur J Med Chem 37:475–486
Galvez J, Garcia-Domenech R, Gomez-Lechon MJ, astell JV (1996) Bioorg Med Chem Lett 6:2301–2306
Via DL, Gia O, Viola G, Bertoloni G, Santana L, Uriarte E (1998) Il Farmaco 53:638–644
Gia O, Anselmo A, Conconi MT, Antonello C, Uriarte E, Caffieri S (1996) J Med Chem 39:4489–4496
Via DL, Gia O, Magno MS, Santana L, Teijeira M, Uriarte E (1999) J Med Chem 42:4405–4413
Fejzo J, Lepre ChA, Peng WJ, Bemis WG, Ajay MAM, Moore MJ (1999) Chem Biol 6:755–769
Gramatica P, Corradi M, Consonni V (2000) Chemosphere 41:763–777
Estrada E, Uriarte E, Montero A, Teijeira M, Santana L, De Clercq E (2000) J Med Chem 43:163–166
Frank IE, Todeschini R (1994) The data analysis handbook. Elsevier, Amsterdam
Gia O, Uriarte E, Zagotto G, Baccichetti F, Antonello C, Marciani-Magno S (1992) J Photochem Photobiol B: Biol 14:95–104
Gia O, Via LD, Marciani S, Angelini G, Margonelli A, Rodighiero P (2000) Photochem Photobiol B 56:132–138
Rodighiero P, Pastorini G, Via LD, Gia O, Marciani S (1998) Il Farmaco 53:313–319
Hermann T, Westhof E (2000) Combinatorial Chem High Throughput Screening 3:219–234
Gozalbes R, Gálvez J, García-Domenech R, Derouin F (1999) SAR QSAR Environ Res 10:47–60
Foye WO, Lemke TL, Williams DA (1995) Principles of medicinal chemistry. Williams and Wilkins, Baltimore, Md., pp 896–900
Arabzadeh A, Bathaiee SZ, Farsam H, Amanlou M, Saboury AA, Shockravi A, Moosavi-Movahedi AA (2002) Int J Pharmaceutics 237:47–45
Spicer JA, Swarna GA, Graene JF, Denny FW (2002) Bioorg Med Chem 10:19–29
Dollery C, Boobis A, Rawlins M, Thomas S, Wilkins M (1999) Therapeitic drugs. Churchill Livingston, Edimburgh, pp 102–108
Scott BR, Pathak MA, Mohn GR (1976) Mutat Res 39:29–74
Bridges BA, Mottershead RP (1977) Mutat Res 44:305–312
Ekins S, Boulanger B, Swaan WP, Hucpey AZ (2002) J Comput Aided Mol Des 16:381-401
Acknowledgements
We would like to offer our sincere thanks to the two unknown referees and the editor for their critical opinions about the manuscript, which have significantly contributed to improving its presentation and quality. González DH would like to express his thanks to Dr. Jose Luis Garcia and the Cuban Ministry of Higher Education for partial financial support and help. The same author acknowledges Dr. Kier L. B. (USA) for his kind revision of other work related to our Markovian model, and who suggested several useful ideas to us. We are also indebted to Dr. Estrada (England) for former tutorship (1994–2000) and training in computational chemistry. We are also grateful for a series of lectures given in Cuba by Dr. Gutman I., which introduced us to the study of the theory of information, and to some extent random process in chemistry. Last but not least, we would like to thank Prof. Nicolais Guevara (Mexico), Prof. Israel Queiroz (Cuba) and Dr. Jorge Galvez (Valencia, Spain) for their useful help, and the Xunta the Galicia for providing us with a grant (PR405A2001/65-0).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gonzáles-Díaz, H., Gia, O., Uriarte, E. et al. Markovian chemicals "in silico" design (MARCH-INSIDE), a promising approach for computer-aided molecular design I: discovery of anticancer compounds. J Mol Model 9, 395–407 (2003). https://doi.org/10.1007/s00894-003-0148-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-003-0148-7