Abstract
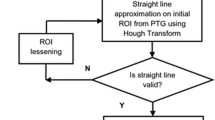
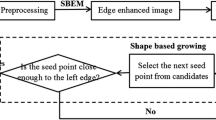
In medio-lateral oblique view of mammogram, pectoral muscle may sometimes affect the detection of breast cancer due to their similar characteristics with abnormal tissues. As a result pectoral muscle should be handled separately while detecting the breast cancer. In this paper, a novel approach for the detection of pectoral muscle using average gradient- and shape-based feature is proposed. The process first approximates the pectoral muscle boundary as a straight line using average gradient-, position-, and shape-based features of the pectoral muscle. Straight line is then tuned to a smooth curve which represents the pectoral margin more accurately. Finally, an enclosed region is generated which represents the pectoral muscle as a segmentation mask. The main advantage of the method is its’ simplicity as well as accuracy. The method is applied on 200 mammographic images consisting 80 randomly selected scanned film images from Mammographic Image Analysis Society (mini-MIAS) database, 80 direct radiography (DR) images, and 40 computed radiography (CR) images from local database. The performance is evaluated based upon the false positive (FP), false negative (FN) pixel percentage, and mean distance closest point (MDCP). Taking all the images into consideration, the average FP and FN pixel percentages are 4.22%, 3.93%, 18.81%, and 6.71%, 6.28%, 5.12% for mini-MIAS, DR, and CR images, respectively. Obtained MDCP values for the same set of database are 3.34, 3.33, and 10.41 respectively. The method is also compared with two well-known pectoral muscle detection techniques and in most of the cases, it outperforms the other two approaches.














Similar content being viewed by others
References
Garcia M, Jemal A, Ward EM, Center MM, Hao Y, Siegel RL, et al: Global cancer facts and figures 2007. American Cancer Society, Atlanta, 2007
American cancer society: Cancer facts and figures 2011. American Cancer Society, Atlanta, 2011
Gupta R, Undrill PE: The use of texture analysis to delineate suspicious masses in mammography. Phys Med Biol 40:835–855, 1995
Hatanaka Y, Hara T, Fujita H, Kasai S, Endo T, Iwase T: Development of an automated method for detecting mammographic masses with a partial loss of region. IEEE Trans Med Imaging 20:1209–1214, 2001
Karssemeijer N: Automated classification of parenchymal patterns in mammograms. Phys Med Biol 43(2):365–378, 1998
Saha PK, Udupa JK, Conant EF, Chakraborty DP, Sullivan D: Breast tissue density quantification via digitized mammograms. IEEE Trans Med Imaging 20:792–803, 2001
Ferrari RJ, Rangayyan RM, Desautels JEL, Frere AF: Segmentation of mammograms: identification of the skin—air boundary, pectoral muscle, and fibro-glandular disc. In IWDM 2000. Proc. 5th Int Workshop Digital Mammography 573, 2001
Kinoshita SK, Azevedo Marques PM, Pereira Jr, RR, Rodrigues JA, Rangayyan RM: Radon-domain detection of the nipple and the pectoral muscle in mammograms. J Digit Imaging 21(1):37–49, 2007
Yam M, Brady M, Highnam R, Behrenbruch C, English R, Kita Y: Three-dimensional reconstruction of microcalcification clusters from two mammographic views. IEEE Trans Med Imaging 20(6):479–489, 2001
Ferrari RJ, Rangayyan RM, Desautels JEL, Borges RA, Frere AF: Automatic identification of the pectoral muscle in mammograms. IEEE Trans Med Imaging 23(2):232–245, 2004
Kwok SM, Chandrasekhar R, Attikiouzel Y, Rickard MT: Automatic pectoral muscle segmentation on mediolateral oblique view mammograms. IEEE Trans Med Imaging 23(9):1129–1140, 2004
Bajger M, Ma F, Bottema MJ. Minimum spanning trees and active contours for identification of the pectoral muscle in screening mammograms. In: Digital Image Computing: Techniques and Applications conference. 47–53, 2005
Ma F, Bajger M, Slavotinek JP, Bottema MJ: Two graph theory based methods for identifying the pectoral muscle in mammograms. Pattern Recogn 40(9):2592–2602, 2007
Wang L, Zhu M, Deng L, Yuan X: Automatic pectoral muscle boundary detection in mammograms based on markov chain and active contour model. Journal of Zhejiang University-SCIENCE C 11(2):111–118, 2010
Cardenosa G: Clinical breast imaging: a patient focused teaching file. Williams & Wilkins, Philadelphia, PA, 2006
Seth S, Mukhopadhyay S, Das P, Bhattacharya P. Shape based breast segmentation in mammograms. IETE Journal of Research, 2011 (in press)
Paredes ESD: Atlas of mammography, 3rd edition. Williams & Wilkins, Philadelphia, 2007
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chakraborty, J., Mukhopadhyay, S., Singla, V. et al. Automatic Detection of Pectoral Muscle Using Average Gradient and Shape Based Feature. J Digit Imaging 25, 387–399 (2012). https://doi.org/10.1007/s10278-011-9421-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-011-9421-y