Abstract
Objectives
To develop orally administrated anti-Helicobacter pylori vaccination, a Lactococcus lactis strain was genetically constructed for fusion expression of H. pylori protective antigens HpaA and Omp22.
Results
The fusion gene of omp22 and hpaA with an adapter encoding three glycines was cloned from a plasmid pMAL-c2x-omp22-hpaA into Escherichia coli MC1061 and L. lactis NZ3900 successively using a shutter vector pNZ8110. Expression of the fusion gene in L. lactis was induced with nisin resulting in production of proteins with molecular weights of 50 and 28 kDa. Both of them were immunoreactive with mouse anti-H. pylori sera as determined via western blotting. Oral vaccination of BALB/c mice using the L. lactis strain carrying pNZ8110-omp22-hpaA elicited significant systematic humoral immune response (P < 0.05).
Conclusions
This is the first report showing that a fusion protein of two H. pylori antigens was efficiently expressed in L. lactis with immunogenicity. This is a considerable step towards H. pylori vaccines.
Similar content being viewed by others
Introduction
Helicobacter pylori infection is closely related to a variety of diseases including gastric adenocarcinoma. Oral vaccination is attractive for control of gastroenteric infection because of safety and effectiveness of the delivery mode (Joan et al. 2016). Lactococcus lactis is a Gram-positive, food-grade bacterium and possesses safety advantage over the attenuated pathogens like Salmonella typhimurium as oral vaccine vectors. In screening of vaccine antigens, the protective effectiveness has drawn more concerns than its safety since the data suggested that side effects of the vaccine candidates were closely related to the adjuvant or delivery vectors instead of the antigens (Anderl and Gerhard 2014). Using L. lactis strains as antigen delivery vehicles, H. pylori urease subunit B (UreB) and Cag7 protein have ever been considered for construction of vaccines. Although these studies showed induced immune responses, they were not sufficient to protect the immunized mice against infection and serve as viable vaccines (Lee et al. 2001; Gu et al. 2009). It is essential therefore to explore more effective vaccine antigens or their combination.
Both H. pylori adhesin A (HpaA) and Omp22 are highly conserved outer membrane proteins. HpaA can provoke cellular maturation of dendritic cells and affect antigen presentation (Flach et al. 2011). Omp22 is structurally related to the OmpA family proteins, most of which are capable of adjusting the innate and adaptive immune responses (Pore et al. 2011). Increasing evidences indicate that HpaA and Omp22 might be promising vaccine antigen candidates (Flach et al. 2011).
In the present study, a fusion gene hpaA–omp22 was cloned into L. lactis NZ3900, the recombinant L. lactis strain was used to vaccinate BALB/c mice, and the serum and mucosal antibody responses were evaluated. This is the first report that a fusion protein of two H. pylori antigens has been efficiently expressed in L. lactis.
Materials and methods
Bacteria, plasmids and animals
Lactococcus lactis NZ3900 and Escherichia coli–L. lactis shuttle vector pNZ8110 were purchased from the NIZO Food Research, Netherlands. The plasmid pMAL-c2x carrying the fusion of omp22 and hpaA genes, designated as pMAL-c2x-omp22-hpaA, was constructed in our previous study (Huang et al. 2013) (Supplementary Table 1). Culture conditions for the bacteria and sources of the materials are as described elsewhere (Chen et al. 2011; Zhang et al. 2009). All animal experiments in this study were approved by the Institutional Review Board at Zhengzhou University, and conducted complying with the ARRIVE guidelines. The SPF 6-week old BALB/c mice were supplied by the Henan Experimental Animal Center.
PCR amplification of the fusion gene of omp22 and hpaA
The plasmid carrying the fusion gene of omp22 and hpaA was extracted from E. coli TB1/pMAL-c2x-omp22-hpaA using alkaline lysis methods, and the omp22-hpaA gene fragment was prepared via PCR using the plasmid pMAL-c2x -omp22-hpaA as template (Huang et al. 2013; Nelson and Fitch 2011). The PCR primers were 5′-GAGCCATGGGCATGAAGAGATCTTCT-3′ (NcoI) and 5′-GCCCTGCAGTTATCGGTTTCTCTTG-3′ (PstI) in sequences. The PCR conditions were as described previously (Huang et al. 2013).
Construction of recombinant plasmids
DNA manipulations, including restrictive endonuclease digestion, ligation, isolation of plasmids from L. lactis, were performed according to the instructions of the suppliers. The omp22-hpaA gene fragment was ligated with pNZ8110 and then used for transformation of E. coli MC1061 and L. lactis NZ3900 successively as described previously (Zhang et al. 2009). L. lactis was transformed by electroporation while E. coli by heat shock (Chen et al. 2011; Zhang et al. 2009).
Preparation and evaluation of mouse anti-H. pylori sera
Helicobacter pylori MEL-Hp27 was cultivated, harvested and lysated ultrasonically as reported by Zhang et al. (2009). The supernatant of cellular lysate mixed with Freund’s adjuvant was used to subcutaneously immunize BALB/c mice for preparation of H. pylori specific antibodies. The immunization of mice and evaluation of serum anti-H. pylori or anti-Omp22-HpaA antibodies were performed as previously described (Zhang et al. 2009).
Omp22-HpaA expression and western blot analysis
The recombinant L. lactis strain NZ3900/pNZ8110-omp22-hpaA was cultivated in GM17 media containing 10 mg chloramphenicol l−1 and induced for expression of the fusion protein Omp22–HpaA under the conditions: inducer nisin 25 μg l−1, OD600 of culture 0.3–0.4 and incubation time 5 h (Chen et al. 2011). Preparation of cellular lysate samples, SDS-PAGE and Western blot analysis were performed using coomassie brilliant blue gel staining and mouse anti-H. pylori sera as described previously (Chen et al. 2011; Zhang et al. 2009). The predicted molecular weight of the recombinant protein was ~50 kDa.
Immunization and sampling
Eighteen BALB/c mice were randomly divided into three groups (n = 6), orally administered with 200 µl recombinant L. lactis NZ3900/pNZ8110-omp22-hpaA (5 × 1011 CFU ml−1), L. lactis NZ3900/pNZ8110 (5 × 1011 CFU ml−1) or PBS four times with intervals of 1 week. The animals were sacrificed 2 weeks after the last bolstering vaccination, and the sera and intestinal fluid were isolated and stored at –20 °C as samples for evaluation of humoral immune responses (Zhang et al. 2009).
Evaluation of humoral immune responses
Escherichia coli TB1/pMAL-c2x-omp22 was employed for expression of Omp22 and maltose binding protein (MBP) as a fusion protein designated as rOmp22, which was purified and used as diagnostic antigen for antibody assay (Huang et al. 2013). The antibody levels in sera and intestinal fluid were evaluated via ELISA. The serum IgG antibody was assayed with diluted serum (1:100) and goat anti-mouse immunoglobulin G labeled with horseradish peroxidase (Cwbiotech, Beijing, China) as the primary and secondary antibodies, respectively. The level of SIgA antibody was measured using intestinal fluid as the primary antibody and polyclonal goat anti-mouse SIgA as secondary antibody (Cellway-Lab, Luoyang, China).
Statistical analysis
The software SPSS17.0 was applied for statistical analysis. The one-way variance analysis of the data was performed to determine difference among the groups. Bonferroni test was used with P value <0.05 as statistically significant.
Results
Amplification of omp22–hpaA
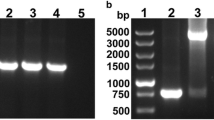
The omp22–hpaA gene fragment was obtained by PCR from the plasmid pMAL-c2x-omp22-hpaA, and the PCR product of omp22–hpaA was estimated to be 1.3 kb.
Construction of a L. lactis strain expressing omp22–hpaA
The omp22–hpaA gene ligated to pNZ8110 was transported to E. coli MC0161 and then L. lactis NZ3900, generating E. coli MC1061/pNZ8110-omp22-hpaA and L. lactis NZ3900/pNZ8110-omp22-hpaA. It was confirmed by gene sequencing that the fusion gene in NZ3900/pNZ8110-omp22-hpaA consisted of the omp22 fragment, a linker (5′-GGTGGAGGC-3′) and the hpaA fragment as shown in Fig. 1. The omp22 and hpaA fragments were corresponding to the published sequences (Genbank No. DQ499023 and DQ353891), respectively, with the terminal codon of omp22 and the start codon of hpaA deleted. Figure 2 shows the identification results of the recombinant L. lactis strain NZ3900/pNZ8110-omp22-hpaA using restrictive enzyme digestion, respectively.
Evaluation of mouse anti-H. pylori sera
The anti-H. pylori sera were collected from mice 1 week post immunization with H. pylori lysate antigens plus Freund’s adjuvant. The double immunodiffusion assays resulted in that the anti-H. pylori sera could recognize the H. pylori cellular sonicate soluble antigens and the Omp22–HpaA fusion protein. There were no co-immunoprecipitation lines between any two of the wells containing H. pylori cellular lysate antigens, Omp22–HpaA, PBS and sera of non immunized mice, respectively. The titer of the antisera reached 1:800 as determined by indirect ELISA.
Expression and immunoreactivity of Omp22–HpaA
To determine whether pNZ8110-omp22-hpaA could drive expression of omp22-hpaA in L. lactis, SDS-PAGE and Western blotting analysis of the cellular lysate was performed. Figure 3 shows the SDS gel visualized by coomassie brilliant blue. There was a much more intense protein band at 28 kDa in the cellular lysate sample of L. lactis NZ3900/pNZ8110-omp22-hpaA, compared with those of the negative controls (Fig. 3). The 28 kDa protein corresponded to the HpaA fragment in molecular weight. The percentage of this protein in cellular extracts of NZ3900/pNZ8110-omp22-hpaA reached 10.2 %. The results of western blot analysis are shown in Fig. 4. The results indicated that the expression product of omp22–hpaA gene in L. lactis consisted of both 28 and 50 kDa proteins, both of which had favorable immunoreactivity with anti-H. pylori sera.
SDS-PAGE analysis of Omp22-HpaA fusion protein expressed in L. lactis transformant. Lane 1 protein molecular weight markers, Lane 2 the cellular lysates of NZ3900/pNZ8110-omp22-hpaA induced using nisin, Lane 3 the cellular lysates of NZ3900/pNZ8110 strain induced using nisin, Lane 4 the cellular lysates of NZ3900 strain induced using nisin
ELISA assays of Omp22-specific antibodies
rOmp22 was obtained with a purity of approximate 90 % by purification from cellular lysate of E. coli TB1/pMAL-c2x-omp22, and used as antigen in ELISA assay of the specific antibody. The sera and intestinal fluid samples were collected from all the mice, and the Omp22-specific antibody was tested, resulting in that serum anti-Omp22 IgG antibody significantly increased in the group vaccinated with NZ3900/pNZ8110-omp22-hpaA, as compared to the groups orally administered with PBS or NZ3900/pNZ8110 (P < 0.05). Nevertheless, there was no significant difference in levels of mucosal SIgA antibodies among all the groups (Table 1).
Discussion
As an expression vector of nisin-controlled gene expression system (NICE) system, the plasmid pNZ8110 had been designed for cytoplasmic or extracellular expression of heteroproteins in suitable hosts, and commonly used for production of bioactive proteins (Zhang et al. 2009; Sánchez et al. 2011). However, a trial conducted earlier in our laboratory did not detect nisin-induced expression of H. pylori UreB both in whole cell lysate and extracellular media of a recombinant L. lactis strain, which had been constructed by insertion of ureB gene between NaeI and XbaI sites in pNZ8110 for transcription fusion of ureB gene and the spusp45 gene fragment under control of nisA promoter. Alteratively, another recombinant L. lactis strain constructed by cloning of ureB gene between the NocI and XbaI in pNZ8110 produced detectable expression of UreB as cytoplasmic protein (Zhang et al. 2009). The reason for this phenomenon is still unclear. Therefore, we cloned the fusion gene into pNZ8110 between the NocI and PstI to make fusion of omp22–hpaA and the nisA promoter. In the fusion gene omp22–hpaA, an oligonucleotide (5′-GGTGGAGGC-3′) encoding a linker peptide had been inserted between the omp22 and hpaA genes. The oligonucleotide encodes a peptide of three glycine residues that helps to keep bioactivity of the proteins in fusion form by enhancing the flexibility of the fusion molecule.
SDS-PAGE analysis showed that only the band at 28 kDa in cellular lysate of NZ3900/pNZ8110-omp22-hpaA was much more intense than that in the controls. Whereas at least two proteins of 28 and 50 kDa, were determined as expression products of omp22–hpaA via western blot assays. The 50 kDa protein is in accordance with the expected expression products of the fusion gene in molecular weight while the 28 kDa protein might be incompletely expressed or decomposed fusion protein. Although only the 28 kDa protein of the two products was distinct in SDS-PAGE pattern, both 50 and 28 kDa bands were clearly visible in Western blots. A possible explanation for this phenomenon is that the 50 kDa product possesses stronger antigenicity or more antigenic epitopes. This study shows that the fusion gene could be efficiently expressed in the recombinant L. lactis strain as cytoplasmic proteins with native antigenicity.
As reported, oral vaccination with purified fusion protein Omp22–HpaA in combination to mLT63 produced protective effect on H. pylori challenged mice with significant decrease in bacterial burden (Huang et al. 2013). Furthermore, a L. lactis delivered vaccine, secreting a multiple epitope antigen, conferred significant humoral immune responses and attenuated H. pylori infection (Li et al. 2014). This study showed that inoculation with NZ3900/pNZ8110-omp22-hpaA strongly up-regulated the serum anti-rOmp22 IgG antibody level, indicating that the fusion protein Omp22–HpaA expressed in L. lactis strain is immunogenic and capable of provoking humoral immune response in mice, and might produce protection against H. pylori challenge. The averaged SIgA level in the group vaccinated with the recombinant L. lactis strain was higher than that in the PBS control group treated with PBS (0.086 ± 0.031 vs. 0.046 ± 0.008), while the difference between the groups was thought as statistically insignificant, a possible explanation for which might be that the sample size designed in this study was not enough large for detection of the difference.
In conclusion, a novel NICE expression system has been constructed in this study for cytoplasmic expression of H. pylori fusion protein Omp22–HpaA in L. lactis. It is the first report that a fusion protein of two H. pylori antigens has been efficiently expressed in L. lactis. This study sets up a model for construction of multivalent oral H. pylori vaccines using L. lactis as a delivery vector, and also provides a hopeful vaccine candidate.
References
Anderl F, Gerhard M (2014) Helicobacter pylori vaccination: is there a path to protection? World J Gastroenterol 20:11939–11949
Chen SY, Zhang RG, Duan GC, Shi JX (2011) Food-grade expression of Helicobacter pylori ureB subunit in Lactococcus lactis and its immunoreactivity. Curr Microbiol 62:1726–1731
Flach CF, Svensson N, Blomquist M, Ekman A, Raghavan S, Holmgren J (2011) A truncated form of HpaA is a promising antigen for use in a vaccine against Helicobacter pylori. Vaccine 29:1235–1241
Gu Q, Song D, Zhu M (2009) Oral vaccination of mice against Helicobacter pylori with recombinant Lactococcus lactis expressing urease subunit B. FEMS Immunol Med Microbiol 56:197–203
Huang X, Xu B, Duan G, Song C (2013) The rOmp22-HpaA fusion protein confers protective immunity against Helicobacter pylori in mice. Curr Microbiol 67:487–492
Joan SS, Pui-Fong J, Song AA, Chang LY, Yusoff K, AbuBakar S, Rahim RA (2016) Oral vaccine of Lactococcus lactis harbouring pandemic H1N1 2009 haemagglutinin1 and nisP anchor fusion protein elevates anti-HA1 sIgA levels in mice. Biotechnol Lett 38:793–799
Lee MH, Roussel Y, Wilks M, Tabaqchali S (2001) Expression of Helicobacter pylori urease subunit B gene in Lactococcus lactis MG1363 and its use as a vaccine delivery system against H. pylori infection in mice. Vaccine 19:3927–3935
Li X, Xing Y, Guo L, Lv X, Song H, Xi T (2014) Oral immunization with recombinant Lactococcus lactis delivering a multi-epitope antigen CTB-UE attenuates Helicobacter pylori infection in mice. Pathog Dis 72:78–86
Nelson MD, Fitch DH (2011) Overlap extension PCR: an efficient method for transgene construction. Methods Mol Biol 772:459–470
Pore D, Mahata N, Pal A, Chakrabarti MK (2011) Outer membrane protein A (OmpA) of Shigella flexneri 2a, induces protective immune response in a mouse model. PLoS One 6:e22663
Sánchez B, López P, González-Rodríguez I, Suárez A, Margolles A, Urdaci MC (2011) A flagellin-producing Lactococcus strain: interactions with mucin and enteropathogens. FEMS Microbiol Lett 318:101–107
Wyszyńska A, Kobierecka P, Bardowski J, Jagusztyn-Krynicka EK (2015) Lactic acid bacteria–20 years exploring their potential as live vectors for mucosal vaccination. Appl Microbiol Biotechnol 99:2967–2977
Zhang XJ, Duan GC, Zhang RG, Fan QT (2009) Optimized expression of Helicobacter pylori ureB gene in Lactococcus lactis NICE system and experimental study on its immunoreactivity. Curr Microbiol 58:308–314
Zhang S, Moise L, Moss SF (2011) H. pylori vaccines: why we still don’t have any. Hum Vaccin 7:1153–1157
Zhang HX, Qiu YY, Zhao YH, Liu XT, Liu M, Yu AL (2014) Immunogenicity of oral vaccination with Lactococcus lactis derived vaccine candidate antigen (UreB) of Helicobacter pylori fused with the human interleukin 2 as adjuvant. Mol Cell Probes 28:25–30
Acknowledgments
We thank the China Postdoctoral Science Foundation (No 200801273) and the Henan Innovation Center of Molecular Diagnosis and Laboratory Medicine (XTCX-2015-ZD2) for financial supports.
Supporting information
Supplementary Table 1—Bacterial strains and plasmids used.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, R., Duan, G., Shi, Q. et al. Construction of a recombinant Lactococcus lactis strain expressing a fusion protein of Omp22 and HpaA from Helicobacter pylori for oral vaccine development. Biotechnol Lett 38, 1911–1916 (2016). https://doi.org/10.1007/s10529-016-2173-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-016-2173-5