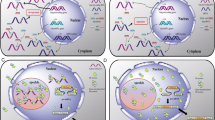
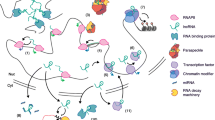
Abstract
The mammalian genome harbors thousands of long noncoding RNA (lncRNA) genes. Recent studies have indicated the involvement of several of these lncRNAs in the regulation of gene expression. lncRNAs play crucial roles in various biological processes ranging from epigenetic gene regulation, transcriptional control, to post-transcriptional regulation. lncRNAs are localized in various subcellular compartments, and major proportion of these are retained in the cell nucleus and could be broadly classified as nuclear-retained lncRNAs (nrRNAs). Based on the identified functions, members of the nrRNAs execute diverse roles, including providing architectural support to the hierarchical subnuclear organization and influencing the recruitment of chromatin modifier factors to specific chromatin sites. In this review, we will summarize the recently described roles of mammalian nrRNAs in controlling gene expression by influencing chromatin organization, transcription, pre-mRNA processing, nuclear organization, and their involvement in disease.


Similar content being viewed by others
Abbreviations
- ANRIL:
-
Antisense nocoding RNA in the INK4 locus
- AR:
-
Androgen receptor
- A-to-I:
-
Adenine to inosine
- CCAT2:
-
Colon cancer-associated transcript 2
- CEBPA:
-
CCAAT/enhancer binding protein (C/EBP), Alpha
- DBHS:
-
Drosophila behavior human splicing
- DHFR:
-
Dihydrofolate reductase
- ecCEBPA:
-
Extracoding CEBPA
- eRNA:
-
Enhancer RNA
- ERα:
-
Estrogen receptor α
- H3K4me3:
-
Histone 3 lysine 4 trimethylation
- H3K27me3:
-
Histone 3 lysine 27 trimethylation
- HAT:
-
Histone acetyl transferase
- hES:
-
Human embryonic stem cells
- HMT:
-
Histone methyltransferase
- hnRNP-K:
-
Heterogeneous nuclear ribonucleoprotein K
- HOTAIR:
-
HOX transcript antisense RNA
- HOTTIP:
-
HOXA transcript at the distal tip
- Igf2r:
-
Insulin-like growth factor 2 receptor
- IGS:
-
Intergenic spacer
- lncRNA:
-
Long noncoding RNA
- MALAT1:
-
Metastasis-associated lung adenocarcinoma transcript 1
- mES:
-
Mouse embryonic stem cells
- MIAT:
-
Myocardial infarction-associated transcript
- ncRNA-a:
-
Noncoding RNA activator
- nrRNA:
-
Nuclear-retained RNA
- PRC2:
-
Polycomb repressive complex 2
- PWS:
-
Prader–Willi syndrome
- RepA:
-
Repeat A
- Schlap1:
-
Second chromosome locus associated with prostate-1
- TCF7L2:
-
transcription factor 7-like 2
- treRNA:
-
Translational regulatory lncRNA
- Uchl1:
-
Ubiquitin carboxyterminal hydrolase L1
- VHL:
-
von Hippel–Lindau
- XCI:
-
X-chromosome inactivation
- Xi:
-
Inactive X-chromosome
- Xic:
-
X-chromosome inactivation center
- Xist:
-
X-inactive-specific transcript
References
Audas TE, Jacob MD, Lee S (2012) Immobilization of proteins in the nucleolus by ribosomal intergenic spacer noncoding RNA. Mol Cell 45:147–157. doi:10.1016/j.molcel.2011.12.012
Augui S, Nora EP, Heard E (2011) Regulation of X-chromosome inactivation by the X-inactivation centre. Nat Rev Genet 12:429–442. doi:10.1038/nrg2987
Batista PJ, Chang HY (2013) Long noncoding RNAs: cellular address codes in development and disease. Cell 152:1298–1307. doi:10.1016/j.cell.2013.02.012
Beckedorff FC, Ayupe AC, Crocci-Souza R, Amaral MS, Nakaya HI, Soltys DT, Menck CF, Reis EM, Verjovski-Almeida S (2013) The intronic long noncoding RNA ANRASSF1 recruits PRC2 to the RASSF1A promoter, reducing the expression of RASSF1A and increasing cell proliferation. PLoS Genet 9:e1003705. doi:10.1371/journal.pgen.1003705
Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan Z, Zhang MQ, Sedel F, Jourdren L, Coulpier F, Triller A, Spector DL, Bessis A (2010) A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J 29:3082–3093. doi:10.1038/emboj.2010.199
Bernstein E, Allis CD (2005) RNA meets chromatin. Genes Dev 19:1635–1655. doi:10.1101/gad.1324305
Bertani S, Sauer S, Bolotin E, Sauer F (2011) The noncoding RNA Mistral activates Hoxa6 and Hoxa7 expression and stem cell differentiation by recruiting MLL1 to chromatin. Mol Cell 43:1040–1046. doi:10.1016/j.molcel.2011.08.019
Berthet C, Aleem E, Coppola V, Tessarollo L, Kaldis P (2003) Cdk2 knockout mice are viable. Curr Biol 13:1775–1785
Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, Kuehn MS, Taylor CM, Neph S, Koch CM, Asthana S, Malhotra A, Adzhubei I, Greenbaum JA, Andrews RM, Flicek P, Boyle PJ, Cao H, Carter NP, Clelland GK, Davis S, Day N, Dhami P, Dillon SC, Dorschner MO, Fiegler H, Giresi PG, Goldy J, Hawrylycz M, Haydock A, Humbert R, James KD, Johnson BE, Johnson EM, Frum TT, Rosenzweig ER, Karnani N, Lee K, Lefebvre GC, Navas PA, Neri F, Parker SC, Sabo PJ, Sandstrom R, Shafer A, Vetrie D, Weaver M, Wilcox S, Yu M, Collins FS, Dekker J, Lieb JD, Tullius TD, Crawford GE, Sunyaev S, Noble WS, Dunham I, Denoeud F, Reymond A, Kapranov P, Rozowsky J, Zheng D, Castelo R, Frankish A, Harrow J, Ghosh S, Sandelin A, Hofacker IL, Baertsch R, Keefe D, Dike S, Cheng J, Hirsch HA, Sekinger EA, Lagarde J, Abril JF, Shahab A, Flamm C, Fried C, Hackermuller J, Hertel J, Lindemeyer M, Missal K, Tanzer A, Washietl S, Korbel J, Emanuelsson O, Pedersen JS, Holroyd N, Taylor R, Swarbreck D, Matthews N, Dickson MC, Thomas DJ, Weirauch MT, Gilbert J et al (2007) Identification and analysis of functional elements in 1 % of the human genome by the ENCODE pilot project. Nature 447:799–816. doi:10.1038/nature05874
Bond CS, Fox AH (2009) Paraspeckles: nuclear bodies built on long noncoding RNA. J Cell Biol 186:637–644. doi:10.1083/jcb.200906113
Bond AM, Vangompel MJ, Sametsky EA, Clark MF, Savage JC, Disterhoft JF, Kohtz JD (2009) Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat Neurosci 12:1020–1027. doi:10.1038/nn.2371
Bulger M, Groudine M (2011) Functional and mechanistic diversity of distal transcription enhancers. Cell 144:327–339. doi:10.1016/j.cell.2011.01.024
Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE (2010) Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet 6:e1001233. doi:10.1371/journal.pgen.1001233
Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE, Batalov S, Forrest AR, Zavolan M, Davis MJ, Wilming LG, Aidinis V, Allen JE, Ambesi-Impiombato A, Apweiler R, Aturaliya RN, Bailey TL, Bansal M, Baxter L, Beisel KW, Bersano T, Bono H, Chalk AM, Chiu KP, Choudhary V, Christoffels A, Clutterbuck DR, Crowe ML, Dalla E, Dalrymple BP, de Bono B, Della GG, di Bernardo D, Down T, Engstrom P, Fagiolini M, Faulkner G, Fletcher CF, Fukushima T, Furuno M, Futaki S, Gariboldi M, Georgii-Hemming P, Gingeras TR, Gojobori T, Green RE, Gustincich S, Harbers M, Hayashi Y, Hensch TK, Hirokawa N, Hill D, Huminiecki L, Iacono M, Ikeo K, Iwama A, Ishikawa T, Jakt M, Kanapin A, Katoh M, Kawasawa Y, Kelso J, Kitamura H, Kitano H, Kollias G, Krishnan SP, Kruger A, Kummerfeld SK, Kurochkin IV, Lareau LF, Lazarevic D, Lipovich L, Liu J, Liuni S, McWilliam S, Madan BM, Madera M, Marchionni L, Matsuda H, Matsuzawa S, Miki H, Mignone F, Miyake S, Morris K, Mottagui-Tabar S, Mulder N, Nakano N, Nakauchi H, Ng P, Nilsson R, Nishiguchi S, Nishikawa S et al (2005) The transcriptional landscape of the mammalian genome. Science 309:1559–1563. doi:10.1126/science.1112014
Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, Pesce E, Ferrer I, Collavin L, Santoro C, Forrest AR, Carninci P, Biffo S, Stupka E, Gustincich S (2012) Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 491:454–457. doi:10.1038/nature11508
Chen LL, Carmichael GG (2008) Gene regulation by SINES and inosines: biological consequences of A-to-I editing of Alu element inverted repeats. Cell Cycle 7:3294–3301
Chen LL, Carmichael GG (2009) Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell 35:467–478. doi:10.1016/j.molcel.2009.06.027
Chen LL, Carmichael GG (2010) Decoding the function of nuclear long non-coding RNAs. Curr Opin Cell Biol 22:357–364. doi:10.1016/j.ceb.2010.03.003
Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, Sementchenko V, Piccolboni A, Bekiranov S, Bailey DK, Ganesh M, Ghosh S, Bell I, Gerhard DS, Gingeras TR (2005) Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 308:1149–1154. doi:10.1126/science.1108625
Chu C, Qu K, Zhong FL, Artandi SE, Chang HY (2011) Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell 44:667–678. doi:10.1016/j.molcel.2011.08.027
Clark MB, Amaral PP, Schlesinger FJ, Dinger ME, Taft RJ, Rinn JL, Ponting CP, Stadler PF, Morris KV, Morillon A, Rozowsky JS, Gerstein MB, Wahlestedt C, Hayashizaki Y, Carninci P, Gingeras TR, Mattick JS (2011) The reality of pervasive transcription. PLoS Biol 9:e1000625. doi:10.1371/journal.pbio.1000625, discussion e1001102
Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB (2009) An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 33:717–726. doi:10.1016/j.molcel.2009.01.026
Concepcion CP, Han YC, Mu P, Bonetti C, Yao E, D’Andrea A, Vidigal JA, Maughan WP, Ogrodowski P, Ventura A (2012) Intact p53-dependent responses in miR-34-deficient mice. PLoS Genet 8:e1002797. doi:10.1371/journal.pgen.1002797
Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R (2010) Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A 107:21931–21936. doi:10.1073/pnas.1016071107
De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G (2010) A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol 8:e1000384. doi:10.1371/journal.pbio.1000384
Denoeud F, Kapranov P, Ucla C, Frankish A, Castelo R, Drenkow J, Lagarde J, Alioto T, Manzano C, Chrast J, Dike S, Wyss C, Henrichsen CN, Holroyd N, Dickson MC, Taylor R, Hance Z, Foissac S, Myers RM, Rogers J, Hubbard T, Harrow J, Guigo R, Gingeras TR, Antonarakis SE, Reymond A (2007) Prominent use of distal 5' transcription start sites and discovery of a large number of additional exons in ENCODE regions. Genome Res 17:746–759. doi:10.1101/gr.5660607
Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigo R (2012) The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22:1775–1789. doi:10.1101/gr.132159.111
Di Ruscio A, Ebralidze AK, Benoukraf T, Amabile G, Goff LA, Terragni J, Figueroa ME, De Figueiredo Pontes LL, Alberich-Jorda M, Zhang P, Wu M, D’Alo F, Melnick A, Leone G, Ebralidze KK, Pradhan S, Rinn JL, Tenen DG (2013) DNMT1-interacting RNAs block gene-specific DNA methylation. Nature. doi:10.1038/nature12598
Do JT, Han DW, Gentile L, Sobek-Klocke I, Stehling M, Scholer HR (2008) Enhanced reprogramming of Xist by induced upregulation of Tsix and Dnmt3a. Stem Cells 26:2821–2831. doi:10.1634/stemcells.2008-0482
Dundr M, Misteli T (2010) Biogenesis of nuclear bodies. Cold Spring Harb Perspect Biol 2:a000711. doi:10.1101/cshperspect.a000711
Eissmann M, Gutschner T, Hammerle M, Gunther S, Caudron-Herger M, Gross M, Schirmacher P, Rippe K, Braun T, Zornig M, Diederichs S (2012) Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol 9:1076–1087. doi:10.4161/rna.21089
Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD (2006) The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev 20:1470–1484. doi:10.1101/gad.1416106
Fox AH, Lamond AI (2010) Paraspeckles. Cold Spring Harb Perspect Biol 2:a000687. doi:10.1101/cshperspect.a000687
Fox AH, Bond CS, Lamond AI (2005) P54nrb forms a heterodimer with PSP1 that localizes to paraspeckles in an RNA-dependent manner. Mol Biol Cell 16:5304–5315. doi:10.1091/mbc.E05-06-0587
Gumireddy K, Li A, Yan J, Setoyama T, Johannes GJ, Orom UA, Tchou J, Liu Q, Zhang L, Speicher DW, Calin GA, Huang Q (2013) Identification of a long non-coding RNA-associated RNP complex regulating metastasis at the translational step. EMBO J. doi:10.1038/emboj.2013.188
Guo F, Li Y, Liu Y, Wang J, Li G (2010) Inhibition of metastasis-associated lung adenocarcinoma transcript 1 in CaSki human cervical cancer cells suppresses cell proliferation and invasion. Acta Biochim Biophys Sin (Shanghai) 42:224–229
Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY (2010) Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464:1071–1076. doi:10.1038/nature08975
Guttman M, Rinn JL (2012) Modular regulatory principles of large non-coding RNAs. Nature 482:339–346. doi:10.1038/nature10887
Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES (2009) Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458:223–227. doi:10.1038/nature07672
Hah N, Murakami S, Nagari A, Danko CG, Kraus WL (2013) Enhancer transcripts mark active estrogen receptor binding sites. Genome Res 23:1210–1223. doi:10.1101/gr.152306.112
Hastings ML, Milcarek C, Martincic K, Peterson ML, Munroe SH (1997) Expression of the thyroid hormone receptor gene, erbAalpha, in B lymphocytes: alternative mRNA processing is independent of differentiation but correlates with antisense RNA levels. Nucleic Acids Res 25:4296–4300
Hastings ML, Ingle HA, Lazar MA, Munroe SH (2000) Post-transcriptional regulation of thyroid hormone receptor expression by cis-acting sequences and a naturally occurring antisense RNA. J Biol Chem 275:11507–11513
Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B (2007) Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39:311–318. doi:10.1038/ng1966
Horiuchi T, Aigaki T (2006) Alternative trans-splicing: a novel mode of pre-mRNA processing. Biol Cell 98:135–140. doi:10.1042/BC20050002
Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, Attardi LD, Regev A, Lander ES, Jacks T, Rinn JL (2010) A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142:409–419. doi:10.1016/j.cell.2010.06.040
Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, Kong B, Langerod A, Borresen-Dale AL, Kim SK, van de Vijver M, Sukumar S, Whitfield ML, Kellis M, Xiong Y, Wong DJ, Chang HY (2011) Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet 43:621–629. doi:10.1038/ng.848
Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A (2007) A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics 8:39. doi:10.1186/1471-2164-8-39
Ishii N, Ozaki K, Sato H, Mizuno H, Saito S, Takahashi A, Miyamoto Y, Ikegawa S, Kamatani N, Hori M, Nakamura Y, Tanaka T (2006) Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J Hum Genet 51:1087–1099. doi:10.1007/s10038-006-0070-9
Jeon Y, Lee JT (2011) YY1 tethers Xist RNA to the inactive X nucleation center. Cell 146:119–133. doi:10.1016/j.cell.2011.06.026
Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, Thomas M, Berdel WE, Serve H, Muller-Tidow C (2003) MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 22:8031–8041. doi:10.1038/sj.onc.1206928
Kanduri C (2011) Kcnq1ot1: a chromatin regulatory RNA. Semin Cell Dev Biol 22:343–350. doi:10.1016/j.semcdb.2011.02.020
Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, Ganesh M, Ghosh S, Piccolboni A, Sementchenko V, Tammana H, Gingeras TR (2007) RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316:1484–1488. doi:10.1126/science.1138341
Kapusta A, Kronenberg Z, Lynch VJ, Zhuo X, Ramsay L, Bourque G, Yandell M, Feschotte C (2013) Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet 9:e1003470. doi:10.1371/journal.pgen.1003470
Kawaji H, Severin J, Lizio M, Waterhouse A, Katayama S, Irvine KM, Hume DA, Forrest AR, Suzuki H, Carninci P, Hayashizaki Y, Daub CO (2009) The FANTOM web resource: from mammalian transcriptional landscape to its dynamic regulation. Genome Biol 10:R40. doi:10.1186/gb-2009-10-4-r40
Kay RA, Ellis IR, Jones SJ, Perrier S, Florence MM, Schor AM, Schor SL (2005) The expression of migration stimulating factor, a potent oncofetal cytokine, is uniquely controlled by 3′-untranslated region-dependent nuclear sequestration of its precursor messenger RNA. Cancer Res 65:10742–10749. doi:10.1158/0008-5472.CAN-05-2038
Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL (2009) Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A 106:11667–11672. doi:10.1073/pnas.0904715106
Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B (2005) A high-resolution map of active promoters in the human genome. Nature 436:876–880. doi:10.1038/nature03877
Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, Markenscoff-Papadimitriou E, Kuhl D, Bito H, Worley PF, Kreiman G, Greenberg ME (2010) Widespread transcription at neuronal activity-regulated enhancers. Nature 465:182–187. doi:10.1038/nature09033
Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, Kim S, Safe S (2013) HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene 32:1616–1625. doi:10.1038/onc.2012.193
Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, Miyano S, Mori M (2011) Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res 71:6320–6326. doi:10.1158/0008-5472.CAN-11-1021
Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y (2011) Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene 30:1956–1962. doi:10.1038/onc.2010.568
Koziol MJ, Rinn JL (2010) RNA traffic control of chromatin complexes. Curr Opin Genet Dev 20:142–148. doi:10.1016/j.gde.2010.03.003
Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, Shiekhattar R (2013) Activating RNAs associate with mediator to enhance chromatin architecture and transcription. Nature 494:497–501. doi:10.1038/nature11884
Lam MT, Cho H, Lesch HP, Gosselin D, Heinz S, Tanaka-Oishi Y, Benner C, Kaikkonen MU, Kim AS, Kosaka M, Lee CY, Watt A, Grossman TR, Rosenfeld MG, Evans RM, Glass CK (2013) Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature 498:511–515. doi:10.1038/nature12209
Lee JT (2011) Gracefully ageing at 50, X-chromosome inactivation becomes a paradigm for RNA and chromatin control. Nat Rev Mol Cell Biol 12:815–826. doi:10.1038/nrm3231
Lee JT (2012) Epigenetic regulation by long noncoding RNAs. Science 338:1435–1439. doi:10.1126/science.1231776
Lee JT, Bartolomei MS (2013) X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell 152:1308–1323. doi:10.1016/j.cell.2013.02.016
Lee JT, Davidow LS, Warshawsky D (1999) Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet 21:400–404. doi:10.1038/7734
Lessing D, Anguera MC, Lee JT (2013) X chromosome inactivation and epigenetic responses to cellular reprogramming. Annu Rev Genomics Hum Genet. doi:10.1146/annurev-genom-091212-153530
Li L, Liu B, Wapinski OL, Tsai MC, Qu K, Zhang J, Carlson JC, Lin M, Fang F, Gupta RA, Helms JA, Chang HY (2013a) Targeted disruption of HOTAIR leads to homeotic transformation and gene derepression. Cell Rep. doi:10.1016/j.celrep.2013.09.003
Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, Oh S, Kim HS, Glass CK, Rosenfeld MG (2013b) Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 498:516–520. doi:10.1038/nature12210
Lin R, Maeda S, Liu C, Karin M, Edgington TS (2007) A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene 26:851–858. doi:10.1038/sj.onc.1209846
Lin R, Roychowdhury-Saha M, Black C, Watt AT, Marcusson EG, Freier SM, Edgington TS (2011) Control of RNA processing by a large non-coding RNA over-expressed in carcinomas. FEBS Lett 585:671–676. doi:10.1016/j.febslet.2011.01.030
Ling H, Spizzo R, Atlasi Y, Nicoloso M, Shimizu M, Redis RS, Nishida N, Gafa R, Song J, Guo Z, Ivan C, Barbarotto E, De Vries I, Zhang X, Ferracin M, Churchman M, van Galen JF, Beverloo BH, Shariati M, Haderk F, Estecio MR, Garcia-Manero G, Patijn GA, Gotley DC, Bhardwaj V, Shureiqi I, Sen S, Multani AS, Welsh J, Yamamoto K, Taniguchi I, Song MA, Gallinger S, Casey G, Thibodeau SN, Le Marchand L, Tiirikainen M, Mani SA, Zhang W, Davuluri RV, Mimori K, Mori M, Sieuwerts AM, Martens JW, Tomlinson I, Negrini M, Berindan-Neagoe I, Foekens JA, Hamilton SR, Lanza G, Kopetz S, Fodde R, Calin GA (2013) CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res 23:1446–1461. doi:10.1101/gr.152942.112
Maamar H, Cabili MN, Rinn J, Raj A (2013) linc-HOXA1 is a noncoding RNA that represses Hoxa1 transcription in cis. Genes Dev 27:1260–1271. doi:10.1101/gad.217018.113
Mao YS, Zhang B, Spector DL (2011) Biogenesis and function of nuclear bodies. Trends Genet 27:295–306. doi:10.1016/j.tig.2011.05.006
Marin-Bejar O, Marchese FP, Athie A, Sanchez Y, Gonzalez J, Segura V, Huang L, Moreno I, Navarro A, Monzo M, Garcia-Foncillas J, Rinn JL, Guo S, Huarte M (2013) Pint lincRNA connects the p53 pathway with epigenetic silencing by the Polycomb repressive complex 2. Genome Biol 14:R104. doi:10.1186/gb-2013-14-9-r104
Martianov I, Ramadass A, Serra BA, Chow N, Akoulitchev A (2007) Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature 445:666–670. doi:10.1038/nature05519
Mattick JS, Amaral PP, Dinger ME, Mercer TR, Mehler MF (2009) RNA regulation of epigenetic processes. Bioessays 31:51–59. doi:10.1002/bies.080099
Melo CA, Drost J, Wijchers PJ, van de Werken H, de Wit E, Oude Vrielink JA, Elkon R, Melo SA, Leveille N, Kalluri R, de Laat W, Agami R (2013) eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol Cell 49:524–535. doi:10.1016/j.molcel.2012.11.021
Mercer TR, Mattick JS (2013) Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol 20:300–307. doi:10.1038/nsmb.2480
Mercer TR, Dinger ME, Mattick JS (2009) Long non-coding RNAs: insights into functions. Nat Rev Genet 10:155–159. doi:10.1038/nrg2521
Mercer TR, Qureshi IA, Gokhan S, Dinger ME, Li G, Mattick JS, Mehler MF (2010) Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci 11:14. doi:10.1186/1471-2202-11-14
Michalak P (2006) RNA world—the dark matter of evolutionary genomics. J Evol Biol 19:1768–1774. doi:10.1111/j.1420-9101.2006.01141.x
Mohammad F, Mondal T, Kanduri C (2009) Epigenetics of imprinted long noncoding RNAs. Epigenetics 4:277–286
Mondal T, Rasmussen M, Pandey GK, Isaksson A, Kanduri C (2010) Characterization of the RNA content of chromatin. Genome Res 20:899–907. doi:10.1101/gr.103473.109
Mousavi K, Zare H, Dell’orso S, Grontved L, Gutierrez-Cruz G, Derfoul A, Hager GL, Sartorelli V (2013) eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol Cell 51:606–617. doi:10.1016/j.molcel.2013.07.022
Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P (2008) The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 322:1717–1720. doi:10.1126/science.1163802
Nakagawa S, Ip JY, Shioi G, Tripathi V, Zong X, Hirose T, Prasanth KV (2012) Malat1 is not an essential component of nuclear speckles in mice. RNA 18:1487–1499. doi:10.1261/rna.033217.112
Ng SY, Bogu GK, Soh BS, Stanton LW (2013) The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol Cell 51:349–359. doi:10.1016/j.molcel.2013.07.017
Niinuma T, Suzuki H, Nojima M, Nosho K, Yamamoto H, Takamaru H, Yamamoto E, Maruyama R, Nobuoka T, Miyazaki Y, Nishida T, Bamba T, Kanda T, Ajioka Y, Taguchi T, Okahara S, Takahashi H, Nishida Y, Hosokawa M, Hasegawa T, Tokino T, Hirata K, Imai K, Toyota M, Shinomura Y (2012) Upregulation of miR-196a and HOTAIR drive malignant character in gastrointestinal stromal tumors. Cancer Res 72:1126–1136. doi:10.1158/0008-5472.CAN-11-1803
Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, Guigo R, Shiekhattar R (2010) Long noncoding RNAs with enhancer-like function in human cells. Cell 143:46–58. doi:10.1016/j.cell.2010.09.001
Ortega S, Prieto I, Odajima J, Martin A, Dubus P, Sotillo R, Barbero JL, Malumbres M, Barbacid M (2003) Cyclin-dependent kinase 2 is essential for meiosis but not for mitotic cell division in mice. Nat Genet 35:25–31. doi:10.1038/ng1232
Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C (2008) Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell 32:232–246. doi:10.1016/j.molcel.2008.08.022
Panganiban G, Rubenstein JL (2002) Developmental functions of the Distal-less/Dlx homeobox genes. Development 129:4371–4386
Pasmant E, Sabbagh A, Vidaud M, Bieche I (2011) ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB J 25:444–448. doi:10.1096/fj.10-172452
Passon DM, Lee M, Rackham O, Stanley WA, Sadowska A, Filipovska A, Fox AH, Bond CS (2012) Structure of the heterodimer of human NONO and paraspeckle protein component 1 and analysis of its role in subnuclear body formation. Proc Natl Acad Sci U S A 109:4846–4850. doi:10.1073/pnas.1120792109
Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, Zhang MQ, Spector DL (2005) Regulating gene expression through RNA nuclear retention. Cell 123:249–263. doi:10.1016/j.cell.2005.08.033
Prensner JR, Chinnaiyan AM (2011) The emergence of lncRNAs in cancer biology. Cancer Discov 1:391–407. doi:10.1158/2159-8290.CD-11-0209
Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, Asangani IA, Grasso CS, Kominsky HD, Cao X, Jing X, Wang X, Siddiqui J, Wei JT, Robinson D, Iyer HK, Palanisamy N, Maher CA, Chinnaiyan AM (2011) Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol 29:742–749. doi:10.1038/nbt.1914
Prensner JR, Iyer MK, Sahu A, Asangani IA, Cao Q, Patel L, Vergara IA, Davicioni E, Erho N, Ghadessi M, Jenkins RB, Triche TJ, Malik R, Bedenis R, McGregor N, Ma T, Chen W, Han S, Jing X, Cao X, Wang X, Chandler B, Yan W, Siddiqui J, Kunju LP, Dhanasekaran SM, Pienta KJ, Feng FY, Chinnaiyan AM (2013) The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet. doi:10.1038/ng.2771
Rinn JL, Chang HY (2012) Genome regulation by long noncoding RNAs. Annu Rev Biochem 81:145–166. doi:10.1146/annurev-biochem-051410-092902
Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY (2007) Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129:1311–1323. doi:10.1016/j.cell.2007.05.022
Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T (2009) MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci U S A 106:2525–2530. doi:10.1073/pnas.0807899106
Schmidt LH, Spieker T, Koschmieder S, Schaffers S, Humberg J, Jungen D, Bulk E, Hascher A, Wittmer D, Marra A, Hillejan L, Wiebe K, Berdel WE, Wiewrodt R, Muller-Tidow C (2011) The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J Thorac Oncol 6:1984–1992. doi:10.1097/JTO.0b013e3182307eac
Schorderet P, Duboule D (2011) Structural and functional differences in the long non-coding RNA HOTAIR in mouse and human. PLoS Genet 7:e1002071. doi:10.1371/journal.pgen.1002071
Shete S, Hosking FJ, Robertson LB, Dobbins SE, Sanson M, Malmer B, Simon M, Marie Y, Boisselier B, Delattre JY, Hoang-Xuan K, El Hallani S, Idbaih A, Zelenika D, Andersson U, Henriksson R, Bergenheim AT, Feychting M, Lonn S, Ahlbom A, Schramm J, Linnebank M, Hemminki K, Kumar R, Hepworth SJ, Price A, Armstrong G, Liu Y, Gu X, Yu R, Lau C, Schoemaker M, Muir K, Swerdlow A, Lathrop M, Bondy M, Houlston RS (2009) Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet 41:899–904. doi:10.1038/ng.407
Shevtsov SP, Dundr M (2011) Nucleation of nuclear bodies by RNA. Nat Cell Biol 13:167–173. doi:10.1038/ncb2157
Shiekhattar R (2013) Opening the chromatin by eRNAs. Mol Cell 51:557–558. doi:10.1016/j.molcel.2013.08.033
Singh DK, Rath PC (2012) Long interspersed nuclear elements (LINEs) show tissue-specific, mosaic genome and methylation-unrestricted, widespread expression of noncoding RNAs in somatic tissues of the rat. RNA Biol 9:1380–1396. doi:10.4161/rna.22402
Sleutels F, Zwart R, Barlow DP (2002) The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 415:810–813. doi:10.1038/415810a
Sone M, Hayashi T, Tarui H, Agata K, Takeichi M, Nakagawa S (2007) The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J Cell Sci 120:2498–2506. doi:10.1242/jcs.009357
Spitale RC, Tsai MC, Chang HY (2011) RNA templating the epigenome: long noncoding RNAs as molecular scaffolds. Epigenetics 6:539–543
Stacey SN, Sulem P, Masson G, Gudjonsson SA, Thorleifsson G, Jakobsdottir M, Sigurdsson A, Gudbjartsson DF, Sigurgeirsson B, Benediktsdottir KR, Thorisdottir K, Ragnarsson R, Scherer D, Hemminki K, Rudnai P, Gurzau E, Koppova K, Botella-Estrada R, Soriano V, Juberias P, Saez B, Gilaberte Y, Fuentelsaz V, Corredera C, Grasa M, Hoiom V, Lindblom A, Bonenkamp JJ, van Rossum MM, Aben KK, de Vries E, Santinami M, Di Mauro MG, Maurichi A, Wendt J, Hochleitner P, Pehamberger H, Gudmundsson J, Magnusdottir DN, Gretarsdottir S, Holm H, Steinthorsdottir V, Frigge ML, Blondal T, Saemundsdottir J, Bjarnason H, Kristjansson K, Bjornsdottir G, Okamoto I, Rivoltini L, Rodolfo M, Kiemeney LA, Hansson J, Nagore E, Mayordomo JI, Kumar R, Karagas MR, Nelson HH, Gulcher JR, Rafnar T, Thorsteinsdottir U, Olafsson JH, Kong A, Stefansson K (2009) New common variants affecting susceptibility to basal cell carcinoma. Nat Genet 41:909–914. doi:10.1038/ng.412
Sun S, Del Rosario BC, Szanto A, Ogawa Y, Jeon Y, Lee JT (2013) Jpx RNA activates Xist by evicting CTCF. Cell 153:1537–1551. doi:10.1016/j.cell.2013.05.028
Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL (2009) MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res 19:347–359. doi:10.1101/gr.087775.108
Tano K, Mizuno R, Okada T, Rakwal R, Shibato J, Masuo Y, Ijiri K, Akimitsu N (2010) MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett 584:4575–4580. doi:10.1016/j.febslet.2010.10.008
Terranova R, Yokobayashi S, Stadler MB, Otte AP, van Lohuizen M, Orkin SH, Peters AH (2008) Polycomb group proteins Ezh2 and Rnf2 direct genomic contraction and imprinted repression in early mouse embryos. Dev Cell 15:668–679. doi:10.1016/j.devcel.2008.08.015
Tian D, Sun S, Lee JT (2010) The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell 143:390–403. doi:10.1016/j.cell.2010.09.049
Tripathi V, Prasanth KV (2011) Cell nucleus. Encycl Life Sci. doi:10.1002/9780470015902.a0001337.pub2
Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, Blencowe BJ, Prasanth SG, Prasanth KV (2010) The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 39:925–938. doi:10.1016/j.molcel.2010.08.011
Tripathi V, Shen Z, Chakraborty A, Giri S, Freier SM, Wu X, Zhang Y, Gorospe M, Prasanth SG, Lal A, Prasanth KV (2013) Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet 9:e1003368. doi:10.1371/journal.pgen.1003368
Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY (2010) Long noncoding RNA as modular scaffold of histone modification complexes. Science 329:689–693. doi:10.1126/science.1192002
Tseng JJ, Hsieh YT, Hsu SL, Chou MM (2009) Metastasis associated lung adenocarcinoma transcript 1 is up-regulated in placenta previa increta/percreta and strongly associated with trophoblast-like cell invasion in vitro. Mol Hum Reprod 15:725–731. doi:10.1093/molehr/gap071
Tsuiji H, Yoshimoto R, Hasegawa Y, Furuno M, Yoshida M, Nakagawa S (2011) Competition between a noncoding exon and introns: Gomafu contains tandem UACUAAC repeats and associates with splicing factor-1. Genes Cells 16:479–490. doi:10.1111/j.1365-2443.2011.01502.x
Turnbull C, Ahmed S, Morrison J, Pernet D, Renwick A, Maranian M, Seal S, Ghoussaini M, Hines S, Healey CS, Hughes D, Warren-Perry M, Tapper W, Eccles D, Evans DG, Hooning M, Schutte M, van den Ouweland A, Houlston R, Ross G, Langford C, Pharoah PD, Stratton MR, Dunning AM, Rahman N, Easton DF (2010) Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet 42:504–507. doi:10.1038/ng.586
Wan G, Hu X, Liu Y, Han C, Sood AK, Calin GA, Zhang X, Lu X (2013) A novel non-coding RNA lncRNA-JADE connects DNA damage signalling to histone H4 acetylation. EMBO J. doi:10.1038/emboj.2013.221
Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, Kurokawa R (2008) Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature 454:126–130. doi:10.1038/nature06992
Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, Glass CK, Rosenfeld MG, Fu XD (2011a) Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature 474:390–394. doi:10.1038/nature10006
Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, Wysocka J, Lei M, Dekker J, Helms JA, Chang HY (2011b) A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472:120–124. doi:10.1038/nature09819
White RJ (2011) Transcription by RNA polymerase III: more complex than we thought. Nat Rev Genet 12:459–463. doi:10.1038/nrg3001
Wilusz JE, Sunwoo H, Spector DL (2009) Long noncoding RNAs: functional surprises from the RNA world. Genes Dev 23:1494–1504. doi:10.1101/gad.1800909
Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, Dorrestein PC, Rosenfeld MG (2011a) ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell 147:773–788. doi:10.1016/j.cell.2011.08.054
Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, Zhang F, Zheng SS (2011b) Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol 18:1243–1250. doi:10.1245/s10434-011-1581-y
Yin QF, Yang L, Zhang Y, Xiang JF, Wu YW, Carmichael GG, Chen LL (2012) Long noncoding RNAs with snoRNA ends. Mol Cell 48:219–230. doi:10.1016/j.molcel.2012.07.033
Yoon JH, Abdelmohsen K, Gorospe M (2012a) Posttranscriptional gene regulation by long noncoding RNA. J Mol Biol. doi:10.1016/j.jmb.2012.11.024
Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, Huarte M, Zhan M, Becker KG, Gorospe M (2012b) LincRNA-p21 suppresses target mRNA translation. Mol Cell 47:648–655. doi:10.1016/j.molcel.2012.06.027
Zhang B, Arun G, Mao YS, Lazar Z, Hung G, Bhattacharjee G, Xiao X, Booth CJ, Wu J, Zhang C, Spector DL (2012) The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep 2:111–123. doi:10.1016/j.celrep.2012.06.003
Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT (2008) Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322:750–756. doi:10.1126/science.1163045
Zhao R, Bodnar MS, Spector DL (2009) Nuclear neighborhoods and gene expression. Curr Opin Genet Dev 19:172–179. doi:10.1016/j.gde.2009.02.007
Zong X, Tripathi V, Prasanth KV (2011) RNA splicing control: yet another gene regulatory role for long nuclear noncoding RNAs. RNA Biol 8:968–977. doi:10.4161/rna.8.6.17606
Acknowledgments
We would like to thank Drs. Stephanie Ceman (UIUC), Ashish Lal (NCI, NIH), and Supriya G. Prasanth (UIUC) for critical reading of the manuscript. We would like to thank Dr. Aparna Anantharaman, Dr. Vidisha Tripathi, and Sumanprava Giri for their assistance in RNA-FISH analyses. Research in the KVP laboratory is supported by grants from American Cancer Society (RSG-11-174-01-RMC) and NIH/NIGMS (GM088252).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editors: Brian P. Chadwick, Kristin C. Scott, and Beth A. Sullivan
Rights and permissions
About this article
Cite this article
Singh, D.K., Prasanth, K.V. Functional insights into the role of nuclear-retained long noncoding RNAs in gene expression control in mammalian cells. Chromosome Res 21, 695–711 (2013). https://doi.org/10.1007/s10577-013-9391-7
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-013-9391-7