Abstract
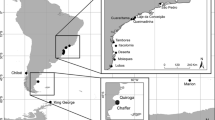
The loggerhead sea turtle, Caretta caretta, is the most common species of sea turtle nesting in Brazil and is listed as endangered by the IUCN. Our study characterizes the genetic structure of loggerheads in Brazil based on mitochondrial DNA control region variability and presents a hypothesis for the colonization of Brazilian rookeries. We analyzed 329 samples from Brazilian rookeries and an oceanic foraging ground, and we compared our results with previously published data for other loggerhead populations. Brazilian rookeries had four haplotypes, none of which have been reported for rookeries outside Brazil. Six haplotypes were found in the foraging aggregation. The presence of the CC-A4 haplotype at all sampled sites and the low nucleotide diversity suggest a common origin for all rookeries, with CC-A4 being the ancestral haplotype of the Brazilian populations. The occurrence of three haplotypes in the foraging aggregation that are known only from rookeries outside of Brazil is consistent with the transoceanic migratory behavior of loggerheads. Our results indicated that the colonization of Brazilian rookeries probably occurred from the southern USA stock. This recent colonization most likely followed a north to south route along the Brazilian coastline, influenced by the Brazilian warm current. Our results further suggest the existence of two genetic population units of loggerheads in Brazil and corroborate natal homing behavior in loggerheads.


Similar content being viewed by others
References
Abreu-Grobois FA, Horrocks J, Formia A, Dutton P, LeRoux R, Vélez Zuazo J, Soares L, Meylan A (2006) New mtDNA D-loop primers which work for a variety of marine turtle species may increase the resolution of mixed stock analysis. In: Frick M, Panagopoulous A, Rees AF, Williams K (eds) Proceedings of the 26th annual symposium on sea turtle biology and conservation. International Sea Turtle Society, Athens, Greece. Available from http://www.iucnmtsg.org/genetics/meth/primers/abreu_grobois_etal_new_dloop_primers.pdf
ACCSTR (2008) Archie Carr center for sea turtle research. Marine turtle DNA sequence patterns. University of Florida. Available from http://www.accstr.ufl.edu/ccmtdna.html. Accessed 15 Jan 2009
Avise JC, Bowen BW, Lamb T, Meylan AB, Bermingham E (1992) Mitochondrial DNA evolution at a turtle’s pace: evidence for low genetic variability and reduced microevolutionary rate in the Testudines. Mol Biol Evol 9:457–473
Birky CW, Maruyama T, Fuerst P (1983) An approach to population and evolutionary genetic theory for genes in mitochondria and chloroplasts, and some results. Genetics 103:513–527
Bjorndal KA, Bolten AB (2008) Annual variation in source contributions to a mixed stock: implications for quantifying connectivity. Mol Ecol 17:2185–2193
Bolten AB (2003) Active swimmers—passive drifters: the oceanic juvenile stage of loggerheads in the Atlantic system. In: Bolten AB, Witherington BE (eds) Loggerhead sea turtles. Smithsonian Institution Press, Washington, DC, pp 63–78
Bolten AB, Bjorndal KA, Martins HR, Dellinger T, Bicoito MJ, Encalada SE, Bowen BW (1998) Transatlantic developmental migrations of loggerhead sea turtles demonstrated by mtDNA sequence analysis. Ecol Appl 8:1–7
Bowen BW (2003) What is a loggerhead turtle? The genetic perspective. In: Bolten AB, Witherington BE (eds) Loggerhead sea turtles. Smithsonian Institution Press, Washington, DC, pp 7–27
Bowen BW, Karl SA (2007) Population genetics and phylogeography of sea turtles. Mol Ecol 16:4886–4907
Bowen BW, Kamezaki N, Limpus CL, Hughes GL, Meylan AB, Avise JC (1994) Global phylogeography of the loggerhead turtle (Caretta caretta) as indicated by mitochondrial DNA haplotypes. Evol Int J Org Evol 48:1820–1828
Bowen BW, Abreu-Grobois FA, Balazs GH, Kamezaki N, Limpus CJ, Ferl RJ (1995) Trans-Pacific migrations of the loggerhead sea turtle demonstrated with mitochondrial DNA markers. Proc Natl Acad Sci USA 92:3731–3734
Bowen BW, Bass AL, Chow S-M, Bostrom M, Bjorndal KA, Bolten AB, Okuyama T, Bolker BM, Epperly S, Lacasella E, Shaver D, Dodd M, Hopkins-Murphy SR, Musick JA, Swingle M, Rankin-Baransky K, Teas W, Witzell WN, Dutton PH (2004) Natal homing in juvenile loggerhead turtles (Caretta caretta). Mol Ecol 13:3797–3808
Bowen BW, Grant WS, Hillis-Starr Z, Shaver DJ, Bjorndal KA, Bolten AB, Bass AL (2007) Mixed-stock analysis reveals the migrations of juvenile hawksbill turtles (Eretmochelys imbricata) in the Caribbean Sea. Mol Ecol 16:49–60
Bugoni L, Krause L, Petry MV (2001) Marine debris and human impacts on sea turtles in southern Brazil. Mar Poll Bull 42:1330–1334
Carr A (1986) Rips, FADS, and little loggerheads. Bioscience 36:92–100
Carr A (1987) New perspectives on the pelagic stage of sea turtles development. Conserv Biol 1:103–121
Carreras C, Pont S, Maffucci F, Pascual M, Barceló A, Bentivegna F, Cardona L, Alegre F, SantFélix M, Fernández G, Aguilar A (2006) Genetic structuring of immature loggerhead sea turtles (Caretta caretta) in the Mediterranean Sea reflects water circulation patterns. Mar Biol 149:1269–1279
Castelloe J, Templeton AR (1994) Root probabilities for intraspecific gene trees under neutral coalescent theory. Mol Phyl Evol 3:102–113
Clement M, Posada D, Crandall K (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9:1657–1659
Damato ME, Corach D (1996) Genetic diversity of populations of the freshwater shrimp Macrobrachium borelli (Cardidea, Palaemonidae) evaluated by RAPD analysis. J Crustac Biol 16:650–655
Encalada SE, Bjorndal KA, Bolten AB, Zurita JC, Schroeder B, Possardt E, Sears CJ, Bowen BW (1998) Population structure of loggerhead turtle (Caretta caretta) nesting colonies in the Atlantic and Mediterranean as inferred from mitochondrial DNA control region sequences. Mar Biol 130:567–575
Excoffier L, Slatkin M (1995) Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol Biol Evol 12:921–927
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes—application to human mitochondrial DNA restriction data. Genetics 131:479–491
Excoffier L, Laval G, Schneider S (2006) ARLEQUIN version 3.01: an integrated software package for population genetics data analysis. University of Bern, Institute of Zoology, Switzerland. Available from http://cmpg.unibe.ch/software/arlequin3
Fu YX (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147:915–925
Gelman A, Rubin DB (1992) Inference from iterative simulation using multiple sequences. Stat Sci 7:457–511
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Harpending RC (1994) Signature of ancient population growth in a low-resolution mitochondrial DNA mismatch distribution. Hum Biol 66:591–600
Harpending HC, Batzer MA, Gurven M, Jorde LB, Rogers AL, Sherry ST (1998) Genetic traces of ancient demography. Proc Natl Acad Sci USA 95:1961–1967
Huang X, Madan A (1999) CAP3: a DNA sequence assembly program. Genome Res 9:868–877
IUCN (2008) International union for the conservation of nature and natural resources. 2008 IUCN red list of threatened species. Available from http://www.iucnredlist.org/. Accessed 15 Jan 2009
Kotas JE, dos Santos S, de Azevedo VG, Gallo BM, Barata PC (2004) Incidental capture of loggerhead (Caretta caretta) and leatherback (Dermochelys coriacea) sea turtles by the pelagic longline fishery off southern Brazil. Fish Bull 102:93–399
Laurent L, Casale P, Bradai MN, Godley BJ, Gerosa G, Broderick AC, Schroth W, Schierwater B, Levy AM, Freggi D, Abd El-Mawla EM, Hadoud DA, Gomati HE, Domingo M, Hadjichristophorou M, Kornaraky L, Demirayak K, Gautier CH (1998) Molecular resolution of marine turtle stock composition in fishery bycatch: a case study in the Mediterranean. Mol Ecol 7:1529–1542
Limpus CJ, Miller JD, Parmenter CJ, Reimer D, McLachland N, Webb R (1992) Migration of green (Chelonia mydas) and loggerhead (Caretta caretta) turtles to and from eastern Australian rookeries. Wildl Res 19:347–358
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
Marcovaldi MA, Chaloupka M (2007) Conservation status of the loggerhead sea turtle in Brazil: an encouraging outlook. Endanger Species Res 3:133–143
Marcovaldi MA, Patri V, Thomé JC (2005) Projeto TAMAR-IBAMA: twenty-five years protecting Brazilian sea turtles through a community-based conservation programme. Marit Stud 3:39–62
Marcovaldi MA, Sales G, Thomé JC, Dias da Silva AC, Gallo BM, Lima EH, Lima EP, Bellini C (2006) Sea turtles and fishery interactions in Brazil: identifying and mitigating potential conflicts. Mar Turtle Newsl 112:4–8
Meylan AB (1982) Sea turtle migration-evidence from tag returns. In: Bjorndal KA (ed) Biology and conservation of sea turtles. Smithsonian Institution Press, Washington, DC, pp 91–100
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Pella J, Masuda M (2001) Bayesian methods for analysis of stock mixtures from genetic characters. Fish Bull 99:151–167
Pritchard PCH, Trebbau P (1984) The turtles of Venezuela. Contributions in herpetology 2, society for the study of amphibians and reptiles. Fundación de Internados Rurales, Caracas
Raymond M, Rousset F (1995) An exact test for population differentiation. Evol Int J Org Evol 49:1280–1283
Reece JS, Ehrhart LM, Parkinson CL (2006) Mixed stock analysis of juvenile loggerheads (Caretta caretta) in Indian River Lagoon, Florida: implications for conservation planning. Cons Gen 7:345–352
Reis EC (2008) Caracterização genética e filogeografia de populações de tartarugas marinhas da espécie Caretta caretta (Linnaeus, 1758) no litoral brasileiro. M.Sc. thesis, Universidade do Estado do Rio de Janeiro, Rio de Janeiro
Rogers AR, Harpending H (1992) Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol 9:552–569
Sales G, Giffoni BG, Barata PCR (2008) Incidental catch of sea turtles by the Brazilian pelagic longline fishery. J Mar Biol Assoc UK 88:853–864
Schneider S, Excoffier L (1999) Estimation of demographic parameters from the distribution of pairwise differences when the mutation rates vary among sites: application to human mitochondrial DNA. Genetics 152:1079–1089
Shanker K, Ramadevi J, Choudhury C, Singh L, Aggarwal RK (2004) Phylogeography of olive ridley turtles (Lepidochelys olivacea) on the east coast of India: implications for conservation theory. Mol Ecol 13:1899–1909
Slatkin M (1987) Gene flow and the geographic structure of natural populations. Science 236:787–792
Snover ML (2002) Growth and ontogeny of sea turtles using skeletochronology: methods, validation and application to conservation. Ph.D. thesis, Duke University, Durham
Soto JMR, Serafini TZ, Celini AAO (2003) Beach strandings of sea turtles in the state of Rio Grande do Sul: an indicator of gillnet interaction along the southern Brazilian coast. In: Seminoff JA (ed) Proceedings of the 22nd annual symposium on sea turtle biology and conservation. NOAA Technical Memorandum NMFS-SEFSC-503, p 276
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526
Templeton AR, Crandall KA, Sing CF (1992) A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics 132:619–633
Wright S (1951) The genetical structure of populations. Ann Eugen 15:323–354
Acknowledgments
We are grateful to CENPES/PETROBRAS (Centro de Pesquisas da PETROBRAS) for supporting the “Mamíferos e Quelônios Marinhos” project, which included this study. The Projeto TAMAR-ICMBio staff collected the samples and provided the necessary field assistance. We acknowledge CAPES, PROCIÊNCIA-SR2-UERJ, FAPERJ and CNPq/MCT for fellowships and grants. The present study followed all ethical guidelines and legal requirements of Brazil for sampling an endangered species.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Reis, E.C., Soares, L.S., Vargas, S.M. et al. Genetic composition, population structure and phylogeography of the loggerhead sea turtle: colonization hypothesis for the Brazilian rookeries. Conserv Genet 11, 1467–1477 (2010). https://doi.org/10.1007/s10592-009-9975-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-009-9975-0