Abstract
The European wildcat, Felis silvestris silvestris, serves as a prominent target species for the reconnection of central European forest habitats. Monitoring of this species, however, appears difficult due to its elusive behaviour and the ease of confusion with domestic cats. Recently, evidence for multiple wildcat occurrences outside its known distribution has accumulated in several areas across Central Europe, questioning the validity of available distribution data for this species. Our aim was to assess the fine-scale distribution and genetic status of the wildcat in its central European distribution range. We compiled and analysed genetic samples from roadkills and hundreds of recent hair-trapping surveys and applied phylogenetic and genetic clustering methods to discriminate wild and domestic cats and identify population subdivision. 2220 individuals were confirmed as either wildcat (n = 1792) or domestic cat (n = 342), and the remaining 86 (3.9 %) were identified as hybrids between the two. Remarkably, genetic distinction of domestic cats, wildcats and their hybrids was only possible when taking into account the presence of two highly distinct genetic lineages of wildcats, with a suture zone in central Germany. 44 % of the individual wildcats where sampled outside the previously published distribution. Our analyses confirm a relatively continuous spatial presence of wildcats across large parts of the study area in contrast to previous analyses indicating a highly fragmented distribution. Our results suggest that wildcat conservation and management should take advantage of the higher than previously assumed dispersal potential of wildcats, which may use wildlife corridors very efficiently.
Similar content being viewed by others
Introduction
Precise knowledge of a species’ current distribution forms the basis for all management actions concerning its conservation. The European Union, for instance, has listed over 1000 animal and plant species that require regular monitoring to assess population status (Council Directive 92/43/EEC of 21 May 1992). Unlike in many other regions, ranges of many central European species, such as large terrestrial mammals seem to be rather well known, as there is considerable public interest in their distribution and they often serve as prominent flagship species for nature conservation (Chapron et al. 2014). Therefore, there is a particular focus on the distribution of this group to document and monitor occurrence, range size and population status (Frosch et al. 2014; Kraus et al. 2015; Simon et al. 2005). In Germany, the European wildcat (Felis silvestris silvestris Schreber 1777) has become a primary target species for promoting large, connected and near-natural broad-leaf forests over the past years. It serves as umbrella and flagship species for endangered forest communities in large-scale conservation projects with the aim to reconnect forest patches in a fragmented and heavily used central European landscape (Vogel and Mölich 2009). Radio-telemetric research data from wildcats were previously used to compute habitat models for the wildcat and resistance values of the landscape were modelled using cost-distance analysis (Vogel and Mölich 2013). In combination with the known distribution data (Birlenbach and Klar 2009) broad-leaf forest corridors were planned and will be implemented throughout Germany where known or potential wildcat habitats appear isolated by anthropogenic barriers or unsuitable habitat (Vogel et al. 2009).
Unlike wolves, lynx and brown bears, the European wildcat was never completely eradicated from western central Europe and survived heavy persecution in several low mountain regions, e.g., the Harz Mountains, the Palatinate Forest, Hunsrück, Taunus and Eifel Mountains in Germany and the Ösling region in Luxembourg. Since the second half of the 20th century, regional recovery of the fragmented populations and range expansion was detected, but with unclear evidence for low-density presence or even long-term persistence in many formerly occupied regions (Knapp et al. 2002; Müller-Using 1962; Piechocki 1986; Raimer 2006; Say et al. 2012). Next to Germany and Luxembourg, there are wildcat populations in France (Say et al. 2012) and Switzerland (Nussberger et al. 2014), whereas for Austria, Belgium and the Netherlands only scattered evidence for wildcat persistence can be found (Dekker et al. 2015; Le Proux de la Rivière and Libois 2006; Slotta-Bachmayr and Friembichler 2010).
Interestingly, precise knowledge on wildcat distribution was lacking until recently, due to its elusive nature and the fact that the species is morphologically similar to wild-coloured domestic cats (Krüger et al. 2009; Müller 2011). As wildcats need to be monitored regularly by EU law (European Council Directive 92/43/EEC, Appendix IV), funds need to be allocated for this purpose. While traditional wildcat monitoring data is based on direct sightings, expert questionnaires, live trapping and roadkill collections (Birlenbach and Klar 2009; Oliveira et al. 2008; Say et al. 2012; Simon et al. 2005), a recent noninvasive monitoring approach using hairs collected with so-called lure stick hair traps has been established allowing for standardized large scale wildcat assessments (Hupe and Simon 2007; Steyer et al. 2013). The lure stick method enables a DNA-based distinction between wild and domestic cat, which promises to solve the long-standing difficulty of safe discrimination under field conditions and the virtual impossibility to safely identify hybrids even under the presence of fresh roadkill material or live-trapped cats (Daniels et al. 1998; Eichholzer 2010; Krüger et al. 2009). As hybridisation with the omnipresent domestic cat was identified as a major threat to the scattered wildcat populations in Europe (Beaumont et al. 2001; Devillard et al. 2014; Nussberger et al. 2014; O’Brien et al. 2009; Pierpaoli et al. 2003), the safe discrimination of wild and domestic cats and their hybrids poses another major advantage of lure stick-based monitoring compared to traditional survey methods.
These advantages along with above mentioned legal monitoring requirements recently led to the rapid spread of genetic wildcat monitoring in the German-speaking countries (Hartmann et al. 2013; Kéry et al. 2011). In Germany and Luxembourg >500 lure stick projects were initiated since 2007 (own data). These surveys were carried out by >100 different institutions, such as regional or state environmental authorities, forestry agencies, NGOs, scientific institutions, but also by interested citizens and schools and showed a broad variety of different sampling schemes in terms of inspection intervals, sampling period, and distance between lure sticks and training of staff. Only few large-scale systematic wildcat surveys (e.g. “Wildcat Leap Project”, Friends of the Earth Germany BUND) used systematic spatial sampling grids. In contrast to this project, which comprises sampling grids across the species’ distribution in Germany for three consecutive years (Vogel and Mölich 2013), the majority of surveys were restricted to small spatial and temporal scales. In addition, we obtained genetic samples from roadkill monitoring including morphometric results and cats captured in the framework of various different telemetry studies.
Here we present a first synopsis of the ongoing large-scale wildcat survey conducted between 2007 and 2013 based on over 6000 samples. We analysed hair samples in the frame of the above mentioned wildcat surveys and collected additional genetic samples from roadkills as well as samples from cats captured in the frame of telemetry studies. Our aims were to (i) describe the current distribution of wildcats in the study range solely based on genetically confirmed survey data, (ii) to reveal large-scale patterns of population structure in the species, and (iii) to obtain a first comprehensive estimate of the genetic integrity of wildcats in the study area based on extensive sampling.
To document the advantages of large scale noninvasive genetic assessment of wildlife as a prime example and in general we provide detailed information on the specificity of lure stick sampling and demonstrate the feasibility of the method by highlighting success rates of various sample sources and analyses from over 6000 samples.
Methods
Sample collection and DNA extraction
Overall, 6019 samples were collected in Germany and Luxembourg, with 97 % of the samples collected between 2007 and 2013 (Table 1). Hair samples were collected primarily with the lure stick method (Hupe and Simon 2007; Steyer et al. 2013) using valerian as an attractant according to the guidelines provided by Hartmann et al. (2013). Lure sticks were predominantly placed in forest habitats and over 95 % of the samples were collected in the mating period of wildcats between December and May. Hair samples were usually stored in filter papers in plastic bags filled with silica gel to keep samples dry. However, due to the fact that samples were collected by multiple collectors in the frame of various surveys, a wide range of other collection methods were applied as well. Sample collection of roadkills was either opportunistically or systematically performed in regional and supra-regional surveys in most German federal states and Luxembourg (Simon et al. 2011; Steeb 2015). Morphometric analyses of carcasses were performed by experts (FM, MK, SS) and were based on morphological (e.g. intestine length, cranial volume) and partially on pelage (e.g. tail bands) characteristics following Krüger et al. (2009) and Müller (2011). All tissue samples were stored in 96 % non-denatured ethanol, hair samples of roadkills were stored as described above. Genetic samples of captured cats, like blood, hair or saliva, were obtained as by-products of telemetry studies or routine analyses of veterinarians in compliance with the respective local and national laws. Cotton swabs with saliva were stored in plastic bags with silica, and blood samples were preserved with EDTA. No animal was sacrificed for the purposes of this study.
Isolation of DNA from hair samples was performed as described in Steyer et al. (2013) in a separate laboratory room dedicated to the pre-PCR handling of noninvasively collected samples (Taberlet et al. 1999). Following the instructions of the manufacturer, the Qiagen Investigator Kit (Hilden, Germany) was used for hair and saliva samples with an additional incubation step at the final elution step for 5 min. Hair samples with more than five hairs with roots (n = 4866) were eluted twice with each 40 µl of ATE buffer, samples with fewer roots were incubated twice with each 20 µl ATE (n = 482). Blood and tissue samples were processed with the Qiagen Blood and Tissue Kit (Qiagen) as recommended by manufacturer´s instructions. Negative controls were run alongside all extractions and PCR reactions to monitor for possible cross contamination.
Mitochondrial DNA analysis
From 5051 samples a Felis-specific part of the mitochondrial control region (110 base pairs) was sequenced with the primers LF4 (Eckert et al. 2010) and H16498 (Kocher et al. 1989) following the protocol in Steyer et al. (2013). Lure stick samples which showed no amplification were optionally sequenced with an additional, less specific mtDNA marker with primers CanidC1 (Paxinos et al. 1997) and HCarn200 (Bidlack et al. 2007), designed for mammal species identification. Sequences were aligned with the ClustalW algorithm (Thompson et al. 1994) in Geneious 7.1.7 (Biomatters). Additionally, we included four previously published sequences (see supplementary Table S1) from GenBank (Benson et al. 2015) to our dataset and created a statistical parsimony network in TCS 1.21 (Clement et al. 2000), treating gaps as a fifth character state. Haplotype and nucleotide diversity were calculated using DNASP 5.10 (Librado and Rozas 2009).
Microsatellite genotyping and analysis
We analysed 4004 cat samples (3212 lure stick/652 roadkill/140 captured cats) with 14 microsatellite markers and a sex marker according to Hartmann et al. (2013). We amplified microsatellites and sex marker in four multiplex reactions (Steyer et al. 2013) and applied a multiple tubes approach with three replicates for hair and saliva samples to account for genotyping errors due to low quality and concentration of template DNA (Navidi et al. 1992). For 80 % of the blood samples and 90 % of the tissue samples at least two replicates were realised to check for consistency. Fragment length analysis was performed on an ABI 3730 DNA Analyzer (Applied Biosystems) using LIZ500 as a size standard and raw data was scored using GeneMarker 2.2 (SoftGenetics). Consensus genotypes for all samples and error rates for allelic drop-out and false alleles for samples with a minimum of eleven loci were calculated with a customised script in R (R Core Team 2014), based on Gimlet (Valière 2002), accepting a heterozygote locus if it was found at least once in the replicates.
Individualisation was performed using the consensus genotype in a customised R script combining genotype information of samples with all relevant information for each sample, such as sampling date, locality, haplotype information, error rates and sex. Substructure among the sampled cats was analysed using the Bayesian clustering algorithm implemented in Structure (Pritchard et al. 2000) with 100,000 MCMC steps after discarding the first 10,000 steps as burn-in, under the admixture model with correlated allele frequencies. No prior information was used. A range of K = 1–21 was tested with ten independent replicates. The results of the ten replicate runs for each value of K were combined using the Greedy algorithm of Clumpp (Jakobsson and Rosenberg 2007). The most likely K values were selected using the Evanno method (Evanno et al. 2005), implemented in Structure Harvester (Earl and vonHoldt 2011). Individuals were assigned to one cluster if their Q value (qi) was qi ≥ 0.8 (Oliveira et al. 2007; Pierpaoli et al. 2003; Witzenberger and Hochkirch 2014). Samples with qi < 0.8 to any cluster and qi < 0.2 to domestic cat clusters were classified as admixed wildcats, and samples with qi > 0.2 to domestic cat clusters and qi < 0.8 to wildcat clusters were classified as potential hybrids between wildcat and domestic cat. Cluster identification was based on morphometrically determined wild and domestic cats which were included in all analyses. Basic population genetic measures such as mean number of alleles per locus (N A), number of private alleles (N P), observed (H O) and expected (H E) heterozygosity and Wright´s fixation index (F IS) were calculated for every subpopulation based on Structure results with GenAlEx 6 (Peakall and Smouse 2006). Deviations from Hardy–Weinberg equilibrium (HWE) were assessed with GenAlEx 6 (Peakall and Smouse 2006) and statistical significance at a nominal threshold of p < 0.05 was evaluated by sequential Bonferroni correction (Holm 1979; Rice 1989). F ST values to measure population differentiation were calculated with Genetix 4.05 (Belkhir et al. 1996) and statistical significance between all population pairs was estimated using 1.000 permutations in Arlequin (Excoffier and Lischer 2010). For the description of population differentiation by genetic variation Factorial correspondence analysis (FCA) using Genetix 4.05 (Belkhir et al. 1996) and Principal Component Analysis (PCA) using adegenet package (Jombart 2008) for R 2.15.3 (R Core Team 2014) were computed. Multilocus genotypes in both FCA and PCA were evaluated with available morphometric data and results from relevant Structure runs.
Wildcat distribution
For Germany (DE), the latest revision in terms of distribution was done by Birlenbach and Klar (2009), who collected wildcat abundance data provided by local and regional experts and classified distribution in two categories: (i) persistent presence based on high abundance and reproduction evidence (18,000 km2) and (ii) single detection of wildcats based on roadkills and genetic data between 1999 and 2009 (Fig. 1c). The single detections, e.g., in the Rhine valley, the Rhön-, the Westerwald-, Kellerwald-, Burgwald-, Rothaar-Mountains and the Egge Hills indicate wildcat presence, but do not allow to draw conclusions concerning the existence of stable populations in these areas. For Luxembourg (LU), wildcat data based on observations, roadkills and genetic monitoring between 2007 and 2012 (Moes 2009; Pir et al. 2011) lead to a point distribution map (Schneider and Sowa 2014). Results of samples analysed in this study which were included in the map from Schneider and Sowa (2014) were compared to the remaining data of the map.
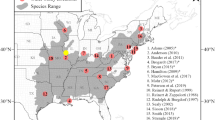
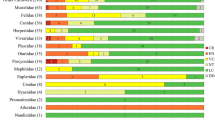
Distribution of cat samples and Structure results for 2220 individuals. a samples per 10 × 10 km grid cell which were genetically confirmed as Felis samples (mtDNA and/or microsatellite analysis; 619 grid cells; n = 4876), b genetic substructuring of cat samples from Germany and Luxembourg using a minimum of eleven microsatellite loci. No prior non-genetic information was used. Shown are at the top the mean likelihood L(K) and standard deviation (SD) per K value and results based on the Evanno method for estimating the number of subpopulations for 2220 cat individuals (K = 1–21) followed by the Structure plots for K = 2–5. Each bar represents a single individual, dashed lines indicate assignment threshold. The colouration corresponds to the estimated proportions of posterior probability assignments of each sample to each cluster, c displays Structure results for K = 3 (except domestic cats) with corresponding colours to b. Admixed individuals between both wildcat clusters are marked orange, whereas samples from potential hybrids of wildcat and domestic cat are displayed as pink rectangles. Shaded in light grey the wildcat distribution map by Birlenbach and Klar from 2009, d Structure results for K = 5 (except domestic cats) and regions of interest highlighted by red numbers. (Color figure online)
For comparison of the data obtained in this study with the previously mentioned distribution maps of wildcats in DE and LU we used 10 × 10 km grids (Annoni et al. 2004). For Luxembourg, the point data was transferred to a total of 27 grid cells, which harboured at least one observation or genetic evidence (grid cells with genetic evidence n = 18). Hence, as sampling density and frequency differed among regions and surveys, the number of samples per grid or individuals per grid only provides a hint at potential wildcat population densities.
Results
Success rates of mtDNA sequencing and general species identification
For all analysed samples the success rate of obtaining a DNA sequence with Felis-specific primers was 79 % (Table 2). A total number of 1.6 % (n = 70) of the lure stick samples showed signs in the raw data for the presence of two or more sequences harbouring a minimum of two cat haplotypes. The markers CanidC1 and HCarn200 were used for most lure stick samples that gave no clear result with LF4 and H16498 primers (n = 897). For 72 % (n = 649) of these samples the amplification was successful. Twenty-three percent (n = 210) of the analysed samples originated from red fox (Vulpes vulpes), and the remaining samples could be assigned to 13 different mammal species (Table S2). The mtDNA success rate increased from 67 % using a single hair from lure sticks to >85 % when applying a minimum of ten hairs, respectively (supplementary Fig. S1). The 3982 cat sequences displayed 29 different haplotypes (KR076400-76428): 21 already published in GenBank (Table S1) and eight haplotypes observed for the first time in this study.
Mitochondrial DNA diversity and phylogeography
No evidence for a clear separation between wild and domestic cats was found based on the mtDNA haplotype network (Fig. 2). The use of alternative network software, i.e., Network (Bandelt et al. 1999) and SplitsTree (Huson and Bryant 2006) lead to similar outcomes. The 29 haplotypes were compared to the microsatellite inference of the Structure K = 3 run (see below and Table S3/Fig. S2). We found 12 haplotypes in 1430 individuals assigned to wildcats (SNG-HP-FS03/-04/-05/-06/-07/-09/-21/-22/-24/-40/-54/-56) with assignment rates from 88 to 100 % (mean = 97 %) and 12 haplotypes (SNG-HP-13/-16/-26/-32/-34/-36/-37/-41/-47/-48/-52/-53) in 248 individuals assigned to domestic cats with assignment rates ranging from 86 to 100 % (mean = 96 %). Four haplotypes (SNG-HP-FS12/-15/-23/-39) found in 21 individuals could not be assigned to domestic or wildcat. For one haplotype (SNG-HP-FS31) no microsatellite data could be obtained. We visually identified three groups in the network that can be mainly assigned to wildcats (a) and domestic cats (b, c) based on microsatellite results (Fig. 2). Haplotypes FS22, FS03 and FS04 were the most common wildcat haplotypes in the dataset and based on spatial data were found across the entire sampling area, while wildcat haplotype FS05 occurs exclusively in the western sampling area, and haplotypes FS07, FS40, FS54 and FS56 only on the western side of the Rhine River. Haplotype FS06 was found only on the eastern side of the Rhine River, with a higher frequency further east. Haplotype FS23 was restricted to the Spessart Mountains, which are located in the central southern study region (Fig. S3). Haplotype diversity calculated based on Structure cluster assignments was 0.768 ± 0.005 for wildcats, 0.824 ± 0.004 for domestic cats and 0.837 ± 0.003 within the entire dataset. Consensus sequences grouping wildcat and domestic cat haplotypes based on Structure K = 3 showed seven fixed nucleotide differences (Fig. S5).
Phylogenetic TCS network based on a 110 base pairs fragment of the mitochondrial control region, representing 33 haplotypes in 1782 cat individuals where mitochondrial and microsatellite data was obtained; pie chart colours indicate proportion of membership to one of the three groups as determined by microsatellite analysis: wildcat (black), domestic cat (grey) and potential hybrid genotypes (white); size of the circles indicate the number of samples analysed, see inset on top left. Haplotypes y1, y3, y4, and y6 were obtained from GenBank and not observed in this study (see Table S1 in electronic supplementary material for accession numbers). For haplotype FS31 no microsatellite data could be obtained. Group A can mainly be assigned to wildcats, group B and C mainly to domestic cats, based on microsatellite analysis using Structure and K = 3 (n = 2220)
Microsatellite genotyping success and individual recapture
Ninety-six percent of roadkill samples showed a minimum of eleven successfully genotyped loci, for captured cats 95 % and for lure stick samples 85 %, respectively (Table 3). Highest genotyping success of lure stick samples was achieved with >20 hairs per extract (89 %; Fig. S4). We excluded 46 lure stick samples which showed more than two alleles at minimally two loci, indicative of mixed DNA traces from two or more individuals. The false allele rate was generally low, and the allelic drop-out rate (ADO) was highest in lure stick samples (23 %; Table 3).
In total, 3471 samples achieved the quality criteria and were used for downstream analyses. Individualisation of samples revealed 2220 individual cats (786 females, 1398 males and 36 animals with unknown sex), 88 from Luxembourg and 2132 from Germany. The majority of the individuals sampled with lure sticks (n = 1467) were detected only once (69 %); 14 % of the individuals were sampled twice, while one male wildcat was resampled 27 times (Table S4). Most resampled individuals were detected again within the first sampling period (80 %) and 15 % 1 year later in the next mating period from December to May. The maximum time span between first and last detection was 1449 days. Nine individuals (seven males and two females) detected by lure sticks were rediscovered as roadkill, with a maximum time span of 1278 days (Tables S4/S5). The mean distance between two detections of the same individual was 3 km. However, most detections were actually closer, with a long tail of the distribution extending to a maximum of 45 km (median = 1.6 km, Q1 = 0.5 km, Q3 = 3.3 km).
The sex marker failed to amplify in 1.5 % of the 3471 samples, with lowest amplification rate in hair samples (98 % amplification success). The sex ratio between different kinds of sample origin was similar: for samples from lure stick (33 % females), roadkill (35 % females) and capture (38 % females).
Broad-scale genetic structure and differentiation between wild and domestic cats
Structure analysis with 2220 cat individuals indicated that the most likely number of genetic clusters is K = 3, with a lower support for K = 2 and K = 3 (Fig. 1b). The subdivision of the dataset in two clusters showed 594 individuals with intermediate assignments to both clusters (Fig. 1b; Table 4). The division into three clusters showed one cluster containing 43 of the 46 morphometrically determined domestic cats and two clusters comprising 247 of the 258 wildcat reference samples (Fig. 1b; Table 4). The two wildcat clusters were largely consistent with a geographic division into a western group (wc_west, n = 838) with samples mostly deriving from areas west of the Rhine river and the Taunus Mountains and a central German group (wc_central, n = 857, Fig. 1c). In total, 92 % of all individuals could be clearly assigned to one of the three clusters with mean individual assignment probabilities within each cluster of qi mean = 0.97 (dc and wc_central) and qi mean = 0.96 (wc_west). Ninety-seven individuals were assigned with intermediate probabilities of qi < 0.8 to both wildcat clusters jointly (wc_admixed, 4.4 %), and 86 individuals (3.9 %) were identified as potential hybrids of domestic cat and wildcat (cross table for all Structure runs see Table S6).
Mean number of alleles per locus across all loci was 11.2 for wildcat cluster west, 9.6 for wildcat cluster central (based on K = 3) and 12.1 for the domestic cats. Most loci were not in HWE (dc: 10 of 14; wc_west: 14 of 14; wc_central: 9 of 14) and F IS values were positive in all three populations, ranging from 0.08-0.12 (Table 4 and Table S7).The domestic cat cluster contained 13 private alleles, the wc_central cluster two and the wc_west cluster eight private alleles, and the combination of both wildcat clusters three additional private alleles (Table S8). All F ST calculations for all population subdivisions were highly significant (Table S9) and for K = 3 highest differentiation was found between domestic cats and wildcat individuals from the central German cluster (F ST = 0.17), and less differentiation between domestic cats and wildcats from the western cluster (F ST = 0.09) and between the two wildcat clusters (F ST = 0.08).
To test for further substructuring of the two obtained wildcat clusters, we split the dataset according to the Structure K = 3 results and performed additional Structure runs for K = 1–11 using a dataset comprising only wildcats that could be either assigned to the western or central cluster with a qi ≥ 0.8. The additional Structure runs for the reduced datasets (data not shown) confirmed the split of the western cluster in two separate clusters and the central cluster in two clusters, as obtained for the full dataset using K = 5. For K = 5 using the full dataset with 2220 individuals, a total of 568 individuals could not be assigned to any cluster (Table 4). The domestic cat cluster contained 320 individuals, which clustered in the domestic cat cluster for K = 3, too. In the K = 5 run both the western and central wildcat population were subdivided (Fig. 1d; Table S6). While subdivision of the western population followed the Rhine river, the central wildcat cluster was subdivided into cluster C with most genotypes originating from the area between the rivers Leine and Weser in the northernmost part of the species´ distribution, whereas wildcat cluster D comprises samples from across remaining central population (Fig. 1d). Compared to K = 3, more loci per population were in HWE and showed lower F IS values. Among all four wildcat clusters, cluster A contained six private alleles, whereas cluster B contained two and cluster C one, respectively (Table S8). Genetic differentiation between the four wildcat clusters were all highly significant, showing highest differentiation for cluster C versus cluster A (F ST = 0.12) and cluster B (F ST = 0.12), and lowest differentiation between cluster A and B (F ST = 0.06) and cluster C and D (F ST = 0.06).
An FCA was performed with 2218 multilocus genotypes (two individuals were removed as outliers) with the two axes explaining 6.82 % of the total genetic variability. The analysis showed three groups (Fig. S6): one group contained the morphometrically determined domestic cats, whereas the other two groups comprise the morphometrically determined wildcats. All but one potential hybrids based on morphometrics were grouped as intermediates between the domestic and wildcat groups. Individual assignments for the Structure results, for K = 3, i.e., the domestic cat group and the two wildcat groups were concordant to the domestic, west and central wildcat clusters, which were separated well. The western wildcat cluster showed a closer proximity to the domestic cat group than the central wildcat cluster. A clear separation of the wildcat cluster A and B from the Structure K = 5 run, but not for Clusters C and D could be identified. Same patterns as described for the FCA were observed for the PCA (Fig. S6).
Geographic pattern and distribution of wildcat detections
A total of 619 grid cells harboured genetic evidence of Felis samples according to mtDNA and/or microsatellite analyses (DE n = 606, LU n = 14, Fig. 1a). Based on the microsatellite results for K = 3 including both wildcat clusters and the admixed individuals between both clusters, 453 grid cells harboured at least one wildcat individual (DE = 440; LU = 14), with 278 grid cells comprising at least two individuals (45 %; DE = 269; LU = 10; Fig. 3; Table S10). The highest number of wildcat individuals per grid cell was obtained in the Taunus low mountain range (58 individuals/grid) and in the Harz Mountains (33 individuals/grid). In Germany, 784 (45 %, Fig. 1c) wildcat individuals were collected outside of the previously assumed persistent presence area when sampling location was compared to the map of Birlenbach and Klar (2009). In Luxembourg, the highest number of wildcats was detected in the northern district of Luxembourg with 22 wildcat individuals per grid. In four additonal cells, compared to the remaining data of the distribution map from Schneider and Sowa (2014), genetic wildcat evidence could be confirmed. In two out of the four additional grid cells observations based on phenotpye had been previously made.
Discussion
Large-scale continuous genetic wildcat assessment
The main aim of this study was to provide an up-to-date assessment of wildcat distribution by means of a large scale genetic survey, providing a solid basis for the ongoing plans to reconnect forest patches in Germany. We are unaware of similar projects or sampling strategies resulting in such sampling densities across large areas for rare, elusive species such as the European wildcat.
The most interesting outcome of this study is the high number of wildcat detections and the relatively continuous spatial presence of the species in parts of the study region, which is in marked contrast to the distribution map published in 2009 (Birlenbach and Klar 2009). Obtaining the results of this study was only possible through the involvement of more than 100 project partners and thousands of local volunteers involved in a variety of local lure stick trapping and roadkill monitoring projects. Although genetics is not a cheap task to do, genetic monitoring of rare, elusive carnivores is in many circumstances cheaper than traditional methods that require more intensive fieldwork (de Groot et al. 2016). When comparing our data to the area described by Birlenbach and Klar (2009) it has to be considered that the latter summarised hard distribution data based on viable populations including reproduction evidence available to this date, whereas anecdotal evidence, such as unconfirmed sightings or single roadkill data was not considered sufficient for an area to be included as wildcat range. In many regions defined as areas with no persistent wildcat presence there was substantial evidence for wildcat occurrence, such as the Westerwald-, the Kellerwald-, Burgwald- and Rhön- Mountains or the Egge Hills (Birlenbach and Klar 2009; Liebelt 2008; Schiefenhövel and Klar 2009; Simon and Hupe 2008; Simon et al. 2010). Our results now confirm the assumed or unclear presence of the species in these and further regions and the sex ratio of individuals sampled outside the previous known range (60 % males, 39 % females, 1 % unknown sex) indicates that this observed pattern is not due to male dispersal only. However, some previously unrecognised wildcat occurrences were detected in this study, such as in the Kottenforst (Hörstermann 2012) or the Arnsberg Forest (Kämpfer et al. 2014).
Our approach did not allow to standardise or randomise sampling across space and time, potentially leading to regional and temporal bias. The high density patches in northern Luxembourg and in the Harz and Taunus Mountains in Germany, for instance, were sampled intensively over the last years, providing only a hint at potential wildcat population densities. For sound population size estimation of wildcat populations using the lure stick method, systematic sampling approaches using grid cells and standardized sampling is crucial. Some habitats and southern regions, for instance, where not considered or only fragmentarily sampled in this study. As an example, recent studies also showed wildcat presence in cultural landscapes with good coverage like hedgerows (Götz 2015; Jerosch and Götz 2015; Simon et al. 2015) and wildcats have been recently detected in several regions across southern Germany (Bavaria: Friedrich 2014; Baden-Württemberg: Streif et al. 2012). Thus, our study likely underestimates the current distribution of the species.
This study raises the question if the multitude of new wildcat detections is due to recent range expansions or the fact that lure stick-based genetic analyses led to a previously unreached monitoring intensity. It seems at least plausible that wildcat presence has been overlooked in several regions with very low wildcat densities (Steyer et al. 2013). Traditional wildcat monitoring is usually strongly based on roadkill surveys (Lang et al. 2006), which requires the awareness of local hunters, conservationists and/or road maintenance staff of the possibility of wildcat presence. Due to the rarity of roadkill findings in low-density areas and the ease of confusion with the omnipresent domestic cat, detection of wildcat presence may be regionally hampered over long time frames. To our experience, however, the initial starting-point for intensive local wildcat assessment is the finding of a putative wildcat roadkill, confirmed by genetic analysis, like in the German federal states Baden-Württemberg, Saxony or the Kellerwald-Edersee National Park (Herdtfelder et al. 2007; Stefen 2011; Steyer et al. 2013). In addition, extensive information campaigns led by the BUND increased the general public awareness for the wildcat as a flagship species for the ongoing plans of creating a biotope network (Vogel and Mölich 2013). As a benefit, numerous local lure stick surveys based on citizen science were started in the last years in areas outside the previously known wildcat distribution.
An alternative explanation to the above-discussed lack of sufficiently hard evidence for the species presence is a rapid ongoing range expansion within just a few years. In the Westerwald Mountains, which are located in the western sampling area, Schiefenhövel and Klar (2009) estimated an average speed of range expansion of 800 m/year. In the northern sampling area, range expansion of wildcats was documented since the 1960s, where roadkills and observations gave first evidence of an expansion trend (Pott-Dorfer and Raimer 2004). For the north-western sampling area, first evidence of range expansion was documented in the 1990s in the Kellerwald Mountains (Simon and Hupe 2008), and for the Burgwald and Rothaar Mountains around the year 2000 (Dietz et al. 2015; Simon et al. 2010). Given this evidence, range expansion might have contributed to the observed pattern, but a combination of range expansion and the difficulty to monitor the species in recently established low-density areas appears as the most plausible explanation here. To answer this complex question additional fine-scale analyses of genetic population structure (Bertorelle et al. 2010; Hartmann et al. 2013; Nater et al. 2015) based on the material and genotype data collected in the frame of this study will be the focus of future work.
Evaluation of technical aspects
The plethora of samples used in this study was not collected by scientists or professional wildlife experts, and many collectors had no previous experience with collecting genetic material. For future lure stick surveys on local and regional scales, a standardised protocol which specifies rules for lure stick placement, study period, inspection intervals and distance between hair traps will allow to compare trapping success rates between different studies and regions. Currently, attempts for such a standardisation on national (Federal Agency for Nature Conservation Germany, BfN) and international scale (EUROWILDCAT consortium) are ongoing. However, the simplicity of the lure stick trapping protocol likely contributed to the high success rates of the genetic analyses which was in the range of previous noninvasive genetic wildcat monitoring studies (Hartmann et al. 2013; Steyer et al. 2013).
The allelic drop-out rate for noninvasive collected samples (ADOmean = 23 %) was slightly higher than reported in other studies (Broquet et al. 2007; Hartmann et al. 2013), while higher rates can be found in several genetic studies based on noninvasive samples (Anile et al. 2014: 25%; Frantz et al. 2003: 27%). These error rates were likely due to detrimental environmental conditions for hairs at the lure sticks, such as UV radiation and humidity, negatively affecting DNA quality (Lindahl 1993). Smith and Wang (2014) analysed the effects of sample size and genotyping errors and demonstrated that there was no substantial change in the mean estimates of genetic variation, differentiation and Structure results in the largest sample set with 100 individuals, even for higher ADO rates than observed in this study. Nevertheless, we are aware of the problems that can be caused by high ADO/FA rates in further downstream analysis, such as overestimation of population size (Lampa et al. 2013).
Genetic diversity and substructure within wildcats
We obtained a clear distinction of wildcats and domestic cats based on Structure and the FCA/PCA analyses. Wildcats were subdivided into a western and a central cluster and showed a level of genetic diversity which was in the range of previous studies on wildcats in Germany (Eckert et al., 2010) and France (Say et al. 2012), which does not indicate strong inbreeding or severe recent bottlenecks due to human impact, like deforestation and fragmentation of suitable habitat. The deviations from HWE in our study are likely due to the Wahlund effect (Wahlund 1928), caused by an underlying subtle substructure over large distances. In several regional studies no significant deviations from HWE could be observed (Hartmann et al. 2013; Würstlin 2013). The western lineage harboured higher heterozygosity levels than the central one, as well as more private microsatellite alleles and haplotypes, which can be explained by possible gene flow with adjacent wildcat populations in France and Belgium, like Hille and Pelz (2000) and Pierpaoli et al. (2003) suggested. In contrast, the central lineage is at the north-eastern distribution edge and does not seem to be demographically connected to eastern European wildcat populations (Hertwig et al. 2009), which might explain the apparently lower grade of genetic diversity. We assume that the high proportion of private alleles and haplotypes observed in the western population are the result of an increased historic effective population size compared to the central population.
The contact zone of the central and western wildcat clusters was observed for the first time in this study and is located in the Rothaar- and Westerwald Mountains. The expansion of both lineages towards each other and the resulting ongoing reunification must be a recent process (Dietz et al. 2015; Simon and Hupe 2008), because we observe a clear assignment of individuals to one of the two wildcat lineages and only a few admixed wildcats between the two populations. Also the low frequency of private haplotypes of both lineages in the suture zone indicates the recent reunification and confirms the higher dispersal rate of male wildcats. The differentiation between the two wildcat clusters obtained in this study is lower than the reported F ST values by Eckert et al. (2010), whereas our sampling is representative of the whole continuous distribution range, and we therefore believe that Eckert et al.’s F ST value is likely over-estimating genetic differentiation due to incomplete sampling (Schwartz and McKelvey 2009). Compared to other European wildcat populations, our observed F ST value is higher than the observed genetic differentiation of two bottlenecked wildcat populations in France (Say et al. 2012), but in the same range of two Italian populations, which showed a divergence time similar to the end of the Last Glacial Maximum (Mattucci et al. 2013). Transferred to our study, the central and western lineage might therefore display the recolonisation from different refugia: the central lineage from the Carpathian/Alpine region, whereas for the western lineage the Iberian Peninsula is the most likely refugia (Sommer and Nadachowski 2006). A detailed analysis combining European wildcat samples from the whole distribution range performed by Mattucci et al. (2016) showed that the European wildcat populations are subdivided into five main biogeographic groups, revealing also the split in two central European populations, with divergence times from the Late Pleistocene. Future studies incorporating samples from all possible refugia have to focus on the hypothesis if the central and western lineage obtained in this study are based on recolonisation from different refugia.
The subdivision of German wildcats into a central cluster and a western cluster was already indicated in three previous studies using much smaller numbers of samples and a non-continuous sampling, which was solely based on carcasses as source for genetic material (Hertwig et al. 2009; Mattucci et al. 2016; Pierpaoli et al. 2003). In contrast to these studies, our study revealed an underlying substructure within the two major clades. Not unexpected, the western clade is divided into two distinct subpopulations by the Rhine River valley. The observed values of genetic differentiation (F ST = 0.06) are similar to that obtained by Hartmann et al. (2013) with the identical genetic marker set (F ST = 0.05). The Rhine valley poses a major barrier to wildcat dispersal, as it combines a large stream as well as dense riparian construction density as well as major traffic infrastructure, such as railways and highways. The subdivision of the central wildcat cluster was not as clear as for the western wildcat cluster and the high number of intermediate individuals, which could not be assigned to one of the two central clusters C and D indicate an underlying cryptic population substructure which might be the result of isolation-by-distance.
The combination of mitochondrial and microsatellite data allowed us to identify haplotypes belonging to one of the three groups (domestic vs. wildcat vs. hybrid) with high assignment probability. The proportion of hybrid individuals detected by microsatellites carrying a wildcat haplotype was quite constant, except for haplotype FS23 (KR076412). It was found only in a small geographic range in the Spessart low mountain range, where over 400 wildcats have been released between 1984 and 2011 (Worel 2009). The released individuals came from a large-scale wildcat breeding program with animals captured in eastern parts of Europe (Worel 2009), which explains both the private haplotype found in that region and the high proportion of not clearly assigned individuals. The fact that haplotype FS23 was not found anywhere else in the study region suggests that this extensive local reintroduction, while being locally effective, did not have large-scale effects within the study region.
Estimation of genetic integrity
The European wildcat is one of five existing subspecies of wildcats and population trends are decreasing mainly due to habitat fragmentation and hybridisation with domestic cats (Driscoll et al. 2011; Yamaguchi et al. 2015). Based on molecular data, domestic cats derived from Near Eastern wildcats (Felis s. lybica) and were introduced to Europe around 2000 years ago (Driscoll et al. 2007; Faure and Kitchener 2009). In the past years, different rates of introgression from domestic cat alleles in the European wildcat genepool were reported, ranging from low (Mattucci et al. 2013; Oliveira et al. 2007) to extremely high rates of domestic cat introgression resulting in the local presence of hybrid swarms and the genetic extinction of regional populations (Beaumont et al. 2001; Pierpaoli et al. 2003). Nevertheless, applied marker systems as well as the number of analysed samples show a great variety among studies concerning hybridisation and this should be kept in mind when comparing hybridisation rates in different regions.
The high conformity of genetic results, which were conducted without any a priori information regarding their potential species identity, compared to the morphometrically analysed cats showed that the marker system used in this study is precise in terms of delimitation between wild and domestic cats. None of the pre-classified domestic cats, which were morphometrically determined, were genetically assigned as a wildcat and vice versa. The hybridisation rate between domestic cats and wildcats was 3.9 % for the entire dataset using a K = 3 and compared to other studies, which used different marker sets and thresholds our results indicate that the study region suffers one of the lowest hybridisation rates in Europe (Lecis et al. 2006; Nussberger et al. 2014; Oliveira et al. 2007; Pastor 2012; Say et al. 2012). In contrast to our results, Hertwig et al. (2009) reported hybridisation rates of 42.9 % in the western German wildcat samples and 4.2 % in samples from central Germany, based on admixture analysis comparable to ours and a K = 2. The strong disagreement between the hybridisation rate estimates of the two studies can be explained by the underlying substructuring into a central and a western wildcat lineage that Hertwig et al. (2009) did not take into account. When using a setting of K = 2 in our admixture analysis we obtained misleading results: 60 % of the samples clearly assigned to the western wildcat cluster in K = 3 exhibited dc/wc hybrid signals and 17 % of west wildcats were assigned to the domestic cat cluster. Even morphometrically predetermined wildcats from the western sampling area were wrongly assigned in K = 2 as hybrids between domestic cat and wildcat (n = 79), or to be pure domestic cats (n = 20). The presence of two wildcat lineages must therefore be taken into account when inferring hybridisation between wildcats and domestic cats, especially in the German wildcat populations. Due to the number of microsatellite markers used in this study and the limited power of hybrid detection beyond the F1 and F2 generation, identification of backcrosses was not possible in this study.
Future studies using recently developed SNP panels for detecting domestic cat introgression (Nussberger et al. 2013; Oliveira et al. 2015) will provide more details about the direction of hybridisation and the amount of first and second generation backcrosses. Considering this, we cannot precisely answer if the relatively high number of hybrid individuals found at the western-most range of our sampling area are true hybrids or might be caused by an underlying population substructure with more westerly wildcat populations. Using a reference set of French and Belgian wildcats and/or the use of newly developed single nucleotide polymorphism markers need to clarify this issue in future studies.
Conservation implication
The reconnection of formerly isolated forest patches using the wildcat as a target species is one of the most prominent large-scale conservation projects in central Europe. The data obtained from years of genetic analyses has already altered official distribution maps of the species (BfN 2013) and the novel range information as well as genetic substructure data can now be added in corridor planning and habitat models. The ongoing natural admixture of the obtained western and central population may be accelerated through the implementation of effective corridors. Based on the low hybridisation rate observed across the study region, corridors will not lead to a loss of genetic integrity by connecting local wildcat populations with vast differences in introgression rates. This low hybridisation also verifies the use of mitochondrial haplotypes to discriminate wild and domestic cats providing a cost-effective rapid tool for detecting wildcat presence. We show that the overarching genetic analysis of noninvasively collected samples across multiple studies and involving as many as possible sources of samples is a feasible way to provide detailed insights into the distribution of an elusive species. As most wildcat studies and conservation action plans are designed and conducted on a regional scale there is a lack of ongoing exchange of information on a central scale for planning conservation projects. The collected genotype data from > 1700 wildcat individuals obtained in this study are currently implemented into a web-based genotype database (https://wildkatzendatenbank.de) with the aim to provide wildlife managers, stakeholders and scientists information concerning local wildcat distribution and hybridisation rates. This implementation of a genetic database ultimately allows for the integration of genetic data into conservation practise, exceeding the current use as a tool for species monitoring in wildlife conservation.
References
Anile S, Ragni B, Randi E, Mattucci F, Rovero F (2014) Wildcat population density on the Etna volcano, Italy: a comparison of density estimation methods. J Zool 293:252–261. doi:10.1111/jzo.12141
Annoni A, Bernard L, Lillethun A, Ihde J, Gallego J (2004) Short proceedings of the 1st European workshop on reference grids. 1st workshop on European reference grids, Ispra, Italy, 2003. JRC-Institute for Environment and Sustainability, Ispra
Bandelt HJ, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48
Beaumont M, Barratt EM, Gottelli D, Kitchener AC, Daniels MJ, Pritchard JK, Bruford MW (2001) Genetic diversity and introgression in the Scottish wildcat. Mol Ecol 10:319–336. doi:10.1046/j.1365-294x.2001.01196.x
Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F (1996) GENETIX 4.05, logiciel sous Windows TM pour la génétique des populations. Laboratoire Génome, Populations, Interactions, Université de Montpellier II, Montpellier (France)
Benson DA, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW (2015) GenBank. Nucleic Acids Res 43:D30–D35. doi:10.1093/nar/gku1216
Bertorelle G, Benazzo A, Mona S (2010) ABC as a flexible framework to estimate demography over space and time: some cons, many pros. Mol Ecol 19:2609–2625. doi:10.1111/j.1365-294X.2010.04690.x
BfN (2013) Kombinierte Vorkommens- und Verbreitungskarte der Pflanzen- und Tierarten der FFH-Richtlinie 1363 Felis silvestris (Wildkatze). Bundesamt für Naturschutz Deutschland (BfN, Federal Agency for Nature Conservation Germany). http://www.bfn.de/0316_nat-bericht_2013-komplett.html. Accessed 01 Mar 2016
Bidlack AL, Reed SE, Palsbøll PJ, Getz WM (2007) Characterization of a western North American carnivore community using PCR-RFLP of cytochrome b obtained from fecal samples. Conserv Genet 8:1511–1513. doi:10.1007/s10592-007-9285-3
Birlenbach K, Klar N (2009) Action Plan for the protection of the European wildcat in Germany—protection concept for a target species of the trans-regional network of forest habitats. Naturschutz und Landschaftsplanung 41:325–332
Broquet T, Menard N, Petit E (2007) Noninvasive population genetics: a review of sample source, diet, fragment length and microsatellite motif effects on amplification success and genotyping error rates. Conserv Genet 8:249–260. doi:10.1007/s10592-006-9146-5
Chapron G et al (2014) Recovery of large carnivores in Europe’s modern human-dominated landscapes. Science 346:1517–1519. doi:10.1126/science.1257553
Clement M, Posada D, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9:1657–1659. doi:10.1046/j.1365-294x.2000.01020.x
Daniels MJ, Balharry D, Hirst D, Kitchener AC, Aspinall RJ (1998) Morphological and pelage characteristics of wild living cats in Scotland: implications for defining the ‘wildcat’. J Zool 244:231–247. doi:10.1111/j.1469-7998.1998.tb00028.x
de Groot GA et al (2016) Decades of population genetic research reveal the need for harmonization of molecular markers: the grey wolf Canis lupus as a case study. Mammal Rev 46:44–59. doi:10.1111/mam.12052
Dekker JAJ, Janssen R, Linnartz L, Brouns A, Mulder JL (2015) Activiteiten van een Wilde kat in Zuid-Limburg Natuurhistorisch Maandblad 104:157–162
Devillard S, Jombart T, Leger F, Pontier D, Say L, Ruette S (2014) How reliable are morphological and anatomical characters to distinguish European wildcats, domestic cats and their hybrids in France? J Zool Syst Evol Res 52:154–162. doi:10.1111/jzs.12049
Dietz M, Bögelsack K, Lang J, Simon O (2015) Kyrill und die Wildkatze. Ergebnisse einer Telemetriestudie im Rothaargebirge. In: Volmer K, Simon O (eds) FELIS Symposium vom 16. bis 17. Oktober 2014 in Gießen.- Der aktuelle Stand der Wildkatzenforschung in Deutschland. Schriften des Arbeitskreis Wildbiologie an der Justus-Liebig-Universität Gießen e. V. Verlag Laufersweiler, Gießen
Driscoll CA et al (2007) The near eastern origin of cat domestication. Science 317:519–523. doi:10.1126/science.1139518
Driscoll C, Yamaguchi N, O’Brien SJ, Macdonald DW (2011) A suite of genetic markers useful in assessing wildcat (Felis silvestris ssp.)—domestic cat (Felis silvestris catus) admixture. J Hered 102:S87–S90. doi:10.1093/jhered/esr047
Earl DA, vonHoldt BM (2011) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361. doi:10.1007/s12686-011-9548-7
Eckert I, Suchentrunk F, Markov G, Hartl GB (2010) Genetic diversity and integrity of German wildcat (Felis silvestris) populations as revealed by microsatellites, allozymes, and mitochondrial DNA sequences. Mamm Biol 75:160–174. doi:10.1016/j.mambio.2009.07.005
Eichholzer A (2010) Testing the applicability of pictures taken by camera-traps for monitoring the European wildcat Felis silvestris silvestris in the Jura Mountains of Switzerland. Master Thesis, University of Zürich, Switzerland
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620. doi:10.1111/j.1365-294X.2005.02553.x
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567. doi:10.1111/j.1755-0998.2010.02847.x
Faure E, Kitchener AC (2009) An archaeological and historical review of the relationships between felids and people. Anthrozoos 22:221–238. doi:10.2752/175303709x457577
Frantz AC, Pope LC, Carpenter PJ, Roper TJ, Wilson GJ, Delahay RJ, Burke T (2003) Reliable microsatellite genotyping of the Eurasian badger (Meles meles) using faecal DNA. Mol Ecol 12:1649–1661. doi:10.1046/j.1365-294X.2003.01848.x
Friedrich M (2014) Wildkatze in Bayern. Bayerische Landesanstalt für Wald und Forstwirtschaft LWF aktuell 102:44–46
Frosch C et al (2014) Noninvasive genetic assessment of brown bear population structure in Bulgarian mountain regions. Mamm Biol 79:268–276. doi:10.1016/j.mambio.2014.04.001
Götz M (2015) Die Säugetierarten der Fauna-Flora-Habitat-Richtlinie im Land Sachsen-Anhalt—Wildkatze (Felis silvestris silvestris Schreber, 1777). Berichte des Landesamtes für Umweltschutz Sachsen-Anhalt 2/2015
Hartmann SA, Steyer K, Kraus RHS, Segelbacher G, Nowak C (2013) Potential barriers to gene flow in the endangered European wildcat (Felis silvestris). Conserv Genet 14:413–426. doi:10.1007/s10592-013-0468-9
Herdtfelder M, Strein M, Suchant R (2007) Wildkatzen am Kaiserstuhl. Naturschutz und Landschaftsplanung 39
Hertwig ST, Schweizer M, Stepanow S, Jungnickel A, Bohle UR, Fischer MS (2009) Regionally high rates of hybridization and introgression in German wildcat populations (Felis silvestris, Carnivora, Felidae). J Zool Syst Evol Res 47:283–297. doi:10.1111/j.1439-0469.2009.00536.x
Hille A, Pelz O (2000) Using microsatellite markers for genetic individualization of European wildcats (Felis silvestris) and domestic cats. Bonn Zool Beitr 49:165–176
Holm S (1979) A simple sequentially rejective multiple test procedure. Scand J Stat 6:65–70
Hörstermann M (2012) Erste Ergebnisse bundesweiter Wildkatzen-Inventur: Wildkatze breitet sich aus - dennoch Bedrohung durch zerschnittene Landschaften. Bund für Umwelt und Naturschutz Deutschland (BUND), Bonn/Berlin
Hupe K, Simon O (2007) Die Lockstockmethode–eine nicht invasive Methode zum Nachweis der Europäischen Wildkatze (Felis s. silvestris). Informationsdienst Naturschutz Niedersachsen 27:66–69
Huson DH, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267. doi:10.1093/molbev/msj030
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806. doi:10.1093/bioinformatics/btm233
Jerosch S, Götz M (2015) Wildkatzen in einer Offenland geprägten Kulturlandschaft– Raum-Zeit-Muster und Habitatwahl. In: Volmer K, Simon O (eds) FELIS Symposium vom 16. bis 17. Oktober 2014 in Gießen.- Der aktuelle Stand der Wildkatzenforschung in Deutschland. Schriften des Arbeitskreis Wildbiologie an der Justus-Liebig-Universität Gießen e. V. Verlag Laufersweiler, Gießen
Jombart T (2008) Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405. doi:10.1093/bioinformatics/btn129
Kämpfer S, Klöckener H, Kriegs JO, Rehage HO, Bub G (2014) Wildkatze Felis silvestris nach über 100 Jahren zurück im Briloner Stadtwald, Hochsauerlandkreis. Nat Heim 74:119–121
Kéry M, Gardner B, Stoeckle T, Weber D, Royle JA (2011) Use of spatial capture-recapture modeling and DNA data to estimate densities of elusive animals. Conserv Biol 25:356–364. doi:10.1111/j.1523-1739.2010.01616.x
Knapp J, Kluth G, Herrmann M (2002) Wildkatzen in Rheinland-Pfalz. Naturschutz bei uns Ministerium für Umwelt und Forsten Rheinland-Pfalz 4:24
Kocher TD, Thomas WK, Meyer A, Edwards SV, Pääbo S, Villablanca FX, Wilson AC (1989) Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers. Proc Natl Acad Sci 86:6196–6200
Kraus RHS et al (2015) A single-nucleotide polymorphism-based approach for rapid and cost-effective genetic wolf monitoring in Europe based on noninvasively collected samples. Mol Ecol Resour 15:295–305. doi:10.1111/1755-0998.12307
Krüger M, Hertwig ST, Jetschke G, Fischer MS (2009) Evaluation of anatomical characters and the question of hybridization with domestic cats in the wildcat population of Thuringia, Germany. J Zool Syst Evol Res 47:268–282. doi:10.1111/j.1439-0469.2009.00537.x
Lampa S, Henle K, Klenke R, Hoehn M, Gruber B (2013) How to overcome genotyping errors in non-invasive genetic mark-recapture population size estimation—a review of available methods illustrated by a case study. J Wildl Manag 77:1490–1511. doi:10.1002/jwmg.604
Lang J, Lang S, Müller F (2006) Hinweise für den Umgang mit Todfunden von Wildkatzen. In: Naturschutz-Akademie Hessen, Bund für Umwelt und Naturschutz Deutschland, Institut für Tierökologie und Naturbildung (eds) Kleine Katzen-Große Räume, Fulda. NZH Verlag Wetzlar, pp 101–106
Le Proux de la Rivière B, Libois R (2006) Sauvage… le chat l’est-il toujours autant qu’on le souhaite ? L’Homme & l’Oiseau 44:162–171
Lecis R, Pierpaoli M, Birò ZS, Szemethy L, Ragni B, Vercillo F, Randi E (2006) Bayesian analyses of admixture in wild and domestic cats (Felis silvestris) using linked microsatellite loci. Mol Ecol 15:119–131. doi:10.1111/j.1365-294X.2005.02812.x
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. doi:10.1093/bioinformatics/btp187
Liebelt R (2008) Neuere Nachweise der Wildkatze (Felis silvestris Schreber 1777) im Egge-Weser-Gebiet Beiträge zur Naturkunde zwischen Egge und Weser 20: 135–143
Lindahl T (1993) Instability and decay of the primary structure of DNA. Nature 362:709–715
Mattucci F et al (2013) Genetic structure of wildcat (Felis silvestris) populations in Italy. Ecol Evol 3:2443–2458. doi:10.1002/ece3.569
Mattucci F, Oliveira R, Lyons LA, Alves PC, Randi E (2016) European wildcat populations are subdivided into five main biogeographic groups: consequences of Pleistocene climate changes or recent anthropogenic fragmentation? Ecol Evol 6:3–22. doi:10.1002/ece3.1815
Moes M (2009) Die Wildkatze in Luxemburg. In: Fremuth W, Jedicke E, Kaphegyi T, Wachendörfer V, Weinzierl H (eds) Zukunft der Wildkatze in Deutschland—Ergebnisse des internationalen Wildkatzen-Symposiums 2008 in Wiesenfelden. Erich Schmidt Verlag, Berlin
Müller F (2011) Körpermerkmale als Unterscheidungskriterien zwischen wildfarbenen Hauskatzen (Felis s. catus) und Wildkatzen (Felis silvestris silvestris, Felidae) aus Mitteleuropa. Beiträge zur Jagd- und Wildforschung 36:359–368
Müller-Using D (1962) Neue Daten zur Wiederausbreitung der Wildkatze in der Bundesrepublik. Z Jagdwiss 8:124–126. doi:10.1007/BF01975030
Nater A et al (2015) Reconstructing the demographic history of orang-utans using approximate Bayesian computation. Mol Ecol 24:310–327. doi:10.1111/mec.13027
Navidi W, Arnheim N, Waterman MS (1992) A multiple-tubes approach for accurate genotyping of very small DNA samples by using PCR: statistical considerations. Am J Hum Genet 50:347–359
Nussberger B, Greminger MP, Grossen C, Keller LF, Wandeler P (2013) Development of SNP markers identifying European wildcats, domestic cats, and their admixed progeny. Mol Ecol Resour 13:447–460. doi:10.1111/1755-0998.12075
Nussberger B, Wandeler P, Weber D, Keller LF (2014) Monitoring introgression in European wildcats in the Swiss Jura. Conserv Genet 15:1219–1230. doi:10.1007/s10592-014-0613-0
O’Brien J, Devillard S, Say L, Vanthomme H, Leger F, Ruette S, Pontier D (2009) Preserving genetic integrity in a hybridising world: are European wildcats (Felis silvestris silvestris) in eastern France distinct from sympatric feral domestic cats? Biodivers Conserv 18:2351–2360. doi:10.1007/s10531-009-9592-8
Oliveira R, Godinho R, Randi E, Ferrand N, Alves PC (2007) Molecular analysis of hybridisation between wild and domestic cats (Felis silvestris) in Portugal: implications for conservation. Conserv Genet 9:1–11
Oliveira R, Godinho R, Randi E, Alves PC (2008) Hybridization versus conservation: are domestic cats threatening the genetic integrity of wildcats (Felis silvestris silvestris) in Iberian Peninsula? Philos Trans R Soc B Biol Sci 363:2953–2961
Oliveira R, Randi E, Mattucci F, Kurushima JD, Lyons LA, Alves PC (2015) Toward a genome-wide approach for detecting hybrids: informative SNPs to detect introgression between domestic cats and European wildcats (Felis silvestris). Heredity 115:195–205. doi:10.1038/hdy.2015.25
Pastor M (2012) Etude du degré d’hybridation entre les populations de chats forestiers (Felis sylvestris sylvestris) et de chats harets (Felis sylvestris catus) au Grand-Duché de Luxembourg. Master thesis, Université de Liège, Belgium
Paxinos E, McIntosh C, Ralls K, Fleischer R (1997) A noninvasive method for distinguishing among canid species: amplification and enzyme restriction of DNA from dung. Mol Ecol 6:483–486. doi:10.1046/j.1365-294X.1997.00206.x
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Piechocki R (1986) Ausbreitung, Verluste, Gewichte und Maße der Wildkatze, Felis silvestris Schreber 1777, in der DDR Hercynia NF 23:125–145
Pierpaoli M et al (2003) Genetic distinction of wildcat (Felis silvestris) populations in Europe, and hybridization with domestic cats in Hungary. Mol Ecol 12:2585–2598
Pir JB, Schauls R, Dietz M, Simon O (2011) Bedeutung von Wildbrücken zur Vernetzung von Wanderkorridoren für die Europäische Wildkatze (Felis silvestris silvestris Schreber, 1777) am Beispiel von Pettingen/Mersch (Luxemburg). Bulletin de la Société des naturalistes luxembourgeois 112:59–71
Pott-Dorfer B, Raimer F (2004) Zur Verbreitung der Wildkatze in Niedersachsen. Informationsdienst Naturschutz Niedersachsen 24:279–281
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
R Core Team (2014) R: a language and environment for statistical computing. R Core Team, Vienna
Raimer F (2006) Die Wildkatzenpopulationen in Hessen und Niedersachsen seit dem 18. Jahrhundert—Verfolgung, Bedrohung, Schutz und Wiederausbreitung. In: Naturschutz-Akademie Hessen, Bund für Umwelt und Naturschutz Deutschland, Institut für Tierökologie und Naturbildung (eds) Kleine Katzen-Große Räume, Fulda. NZH Verlag Wetzlar, pp 69–78
Rice WR (1989) Analyzing tables of statistical tests. Evolution 43:223–225. doi:10.2307/2409177
Say L, Devillard S, Léger F, Pontier D, Ruette S (2012) Distribution and spatial genetic structure of European wildcat in France. Anim Conserv 15:18–27. doi:10.1111/j.1469-1795.2011.00478.x
Schiefenhövel P, Klar N (2009) Die Ausbreitung der Wildkatze (Felis silvestris Schreber, 1777) im Westerwald–eine streng geschützte Art auf dem Vormarsch. Fauna Flora Rheinland-Pfalz 11:941–960
Schneider S, Sowa F (2014) Plan national pour la protection de la nature–Plans d’actions espèces–chat sauvage (Felis silvestris silvestris). Ministère du Développement durable et des Infrastructures—Département de l’environnement, Luxembourg
Schwartz MK, McKelvey KS (2009) Why sampling scheme matters: the effect of sampling scheme on landscape genetic results. Conserv Genet 10:441–452. doi:10.1007/s10592-008-9622-1
Simon O, Hupe K (2008) Nachweis der Wildkatze (Felis silvestris silvestris) im Nationalpark Kellerwald-Edersee und den umgebenden Waldgebieten der Breiten Struth, des Hohen Kellers und des nördlichen Burgwaldes mit Hilfe der Lockstockmethode im Winter 2007/08. Jahrbuch Naturschutz in Hessen 12:18–25
Simon O, Hupe K, Trinzen M (2005) Wildkatze (Felis silvestris, Schreber 1977). In: Doerpinghaus A, Eichen C, Gunnermann H, Leopold P, Neukirchen M, Petermann J, Schröder E (eds) Methoden zur Erfassung von Arten der Anhänge IV und V der Fauna-Flora-Habitat-Richtlinie, Naturschutz und Biologische Vielfalt, vol 20. Landwirtschaftsverlag, Münster, pp 395–402
Simon O, Schütz F, Archinal A (2010) Nachweis der Wildkatze (Felis silvestris silvestris) im Burgwald mit Hilfe der Lockstockmethode im Spätwinter 2009 und 2010. Jahrbuch Naturschutz in Hessen 13:61–66
Simon O, Lang J, Steeb S, Eskens U, Müller F, Volmer K (2011) Relevanz der Totfundanalyse von Wildkatzen für das FFH-Monitoring in Deutschland. Beiträge zur Jagd- und Wildforschung 36:377–387
Simon O et al (2015) Rückkehr auf Samtpfoten. Pirsch Deutscher Landwirtschaftsverlag, Hannover 3/2015
Slotta-Bachmayr L, Friembichler S (2010) Aktionsplan Schutz der Wildkatze in Österreich. The Austrian Federal Ministry of Agriculture, Forestry, Environment and Water Management, Wien
Smith O, Wang J (2014) When can noninvasive samples provide sufficient information in conservation genetics studies? Mol Ecol Resour 14:1011–1023. doi:10.1111/1755-0998.12250
Sommer RS, Nadachowski A (2006) Glacial refugia of mammals in Europe: evidence from fossil records. Mamm Rev 36:251–265. doi:10.1111/j.1365-2907.2006.00093.x
Steeb S (2015) Postmortale Untersuchungen an der Europäischen Wildkatze (Felis silvestris silvestris Schreber, 1777). PhD thesis, Justus-Liebig-Universität Gießen, Gießen, Germany
Stefen C (2011) Erster Wildkatzenfund (Felis silvestris Schreber 1777) im Vogtland, Freistaat Sachsen und im Land Brandenburg. Säugetierkundliche Informationen 7:211–221
Steyer K, Simon O, Kraus RHS, Haase P, Nowak C (2013) Hair trapping with valerian-treated lure sticks as a tool for genetic wildcat monitoring in low-density habitats. Eur J Wildl Res 59:39–46. doi:10.1007/s10344-012-0644-0
Streif S, Kraft S, Veith S, Kohnen A, Suchant R (2012) Die Wildkatze (Felis silvestris) in Baden-Württemberg—Statusbericht zu Monitoring und Forschung. Säugetierkundliche Informationen 45:411–416
Taberlet P, Luikart G, Waits LP (1999) Noninvasive genetic sampling: look before you leap. Trends Ecol Evol 14:323–327
Thompson JD, Higgins DG, Gibson TJ (1994) Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Valière N (2002) GIMLET: a computer program for analysing genetic individual identification data. Mol Ecol Notes 2:377–379. doi:10.1046/j.1471-8286.2002.00228.x-i2
Vogel B, Mölich T (2009) Rettungsnetz Wildkatze—Vorhaben zur Wiederherstellung eines Verbundes geeigneter Waldlebensräume. In: Fremuth W, Jedicke E, Kaphegyi T, Wachendörfer V, Weinzierl H (eds) Zukunft der Wildkatze in Deutschland—Ergebnisse des internationalen Wildkatzen-Symposiums 2008 in Wiesenfelden. Erich Schmidt Verlag, Berlin
Vogel B, Mölich T (2013) Best practices for implementing biotope networks in highly fragmented landscapes: the Safety Net for the European wildcat. Paper presented at the proceedings of the 2013 international conference on ecology and transportation, Scottsdale
Vogel B, Mölich T, Klar N (2009) Der Wildkatzenwegeplan: Ein strategisches Instrument des Naturschutz. Naturschutz und Landschaftsplanung 41:333–340
Wahlund S (1928) Composition of populations and correlation appearances viewed in relation to the studies of inheritance. Hereditas 11:65–106
Witzenberger KA, Hochkirch A (2014) The genetic integrity of the ex situ population of the European wildcat (Felis silvestris silvestris) is seriously threatened by introgression from domestic cats (Felis silvestris catus). PLoS ONE 9:e106083. doi:10.1371/journal.pone.0106083
Worel G (2009) Erfahrungen mit der Wiederansiedlung der Wildkatze in Bayern. In: Fremuth W, Jedicke E, Kaphegyi T, Wachendörfer V, Weinzierl H (eds) Zukunft der Wildkatze in Deutschland—Ergebnisse des internationalen Wildkatzen-Symposiums 2008 in Wiesenfelden. Erich Schmidt Verlag, Berlin
Würstlin S (2013) Potential barriers to gene flow in the wildcat (Felis silvestris silvestris) population in the Upper Rhine Valley. Master thesis, University of Freiburg, Germany
Yamaguchi N, Kitchener A, Driscoll C, Nussberger B (2015) Felis silvestris. The IUCN red list of threatened species 2015. http://dx.doi.org/10.2305/IUCN.UK.2015-2.RLTS.T60354712A50652361.en. Accessed 07 Mar 2016
Acknowledgments
For support in the lab we thank João Barateiro Diogo, Susanne Carl and Annakarina Mundorf. For initial work, providing skills and expertise in earlier phases we are grateful to Peter Haase, Martina Denk, Jan Sauer, Kathrin Theißinger and Tabea Stöckle. Sequencing service was provided by the Biodiversity and Climate Research Institute Frankfurt (BiK-F). Structure calculations were performed on the CSC HPC cluster FUCHS of J. W. Goethe University of Frankfurt, Germany. KS was funded during writing by Karl und Marie Schack-Stiftung. RHSK was funded by grant SAW-2011-SGN-3 of the Leibniz Association (Germany) and financially supported by the ESF funded “ConGenOmics Program”. We acknowledge the funding for several projects by the Bund für Umwelt und Naturschutz (BUND) Germany, the German Federal Agency for Nature Conservation (BfN), Hessen-Forst (FENA), Landesbetrieb Wald und Holz North Rhine-Westphalia, Regionalforstamt Hochstift/Egge (North Rhine-Westphalia), Thüringer Landesanstalt für Umwelt und Geologie (TLUG), OEKOFONDS, Arbeitskreis Wildbiologie an der Justus-Liebig-Universität Gießen e.V. (FELIS-Projekt), Landesjagdabgabe Baden-Württemberg, Musée national d’histoire naturelle du Luxembourg, Ministère du Développement durable et des Infrastructures Département de l’environnement Luxembourg (MDDI-DEPENV) and several other funders. We appreciate the guidance, reviews and referral provided by AXIOS review. The authors would like to apologize to all colleagues and thousands of volunteers whose work we could not mention due to space limitations.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Steyer, K., Kraus, R.H.S., Mölich, T. et al. Large-scale genetic census of an elusive carnivore, the European wildcat (Felis s. silvestris). Conserv Genet 17, 1183–1199 (2016). https://doi.org/10.1007/s10592-016-0853-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-016-0853-2