Abstract
Hybridisation between wild taxa and their domestic congeners is a significant conservation issue. Domestic species frequently outnumber their wild relatives in population size and distribution and may therefore genetically swamp the native species. The European wildcat (Felis silvestris) has been shown to hybridise with domestic cats (Felis catus). Previously suggested spatially divergent introgression levels have not been confirmed on a European scale due to significant differences in the applied methods to assess hybridisation of the European wildcat. We analysed 926 Felis spp. samples from 13 European countries, using a set of 86 selected ancestry-informative SNPs, 14 microsatellites, and ten mitochondrial and Y-chromosome markers to study regional hybridisation and introgression patterns and population differentiation. We detected 51 hybrids (four F1 and 47 F2 or backcrosses) and 521 pure wildcats throughout Europe. The abundance of hybrids varied considerably among studied populations. All samples from Scotland were identified as F2 hybrids or backcrosses, supporting previous findings that the genetic integrity of that wildcat population has been seriously compromised. In other European populations, low to moderate levels of hybridisation were found, with the lowest levels being in Central and Southeast Europe. The occurrence of distinct maternal and paternal markers between wildcat and domestic cat suggests that there were no severe hybridisation episodes in the past. The overall low (< 1%) prevalence of F1 hybrids suggests a low risk of hybridisation for the long-term genetic integrity of the wildcat in most of Europe. However, regionally elevated introgression rates confirm that hybridisation poses a potential threat. We propose regional in-depth monitoring of hybridisation rates to identify factors driving hybridisation so as to develop effective strategies for conservation.
Similar content being viewed by others
Introduction
Hybridisation is a naturally occurring process that leads to contradictory evolutionary outcomes. On one hand, it may decrease biodiversity by threatening species or populations with the loss of genetic diversity or outright extinction by genetic swamping (Rhymer and Simberloff 1996; Allendorf et al. 2001; Seehausen et al. 2008; Todesco et al. 2016). On the other hand, hybridisation can contribute to biodiversity by introducing novel genetic diversity and triggering speciation processes (Mallet 2008; Abbott et al. 2016). Globally, a large proportion of species is assumed to be susceptible to hybridisation with related taxa (Seehausen et al. 2008). Hybridisation occurs when populations that have been reproductively isolated for a certain time eventually come into contact, for instance due to range shifts (Futuyma 2005). Currently, rates of hybridisation may also be on the rise due to human-mediated range expansion of alien taxa, leading to previously impossible hybridisation events between naturally allopatric taxa (Mooney and Cleland 2001; Sakai et al. 2001; Simberloff et al. 2013). Moreover, the introduction of actively managed taxa for husbandry and sport hunting, as in the case of livestock, pets and game, may also enhance opportunities for hybridisation (Todesco et al. 2016). Non-native and, in particular, domestic taxa have been introduced on a global scale and often population sizes are substantially larger compared to those of their wild congeners. In addition to this, traits of domestic taxa have been artificially selected to meet human needs; the introgression of “domestic” genes in wild taxa may thus lead to decreased fitness or to outbreeding depression in wild populations (Todesco et al. 2016). Therefore, studying the distribution and causes of hybridisation between populations of wild and domestic taxa is highly relevant for species conservation. Hybridisation between wild and domestic congeners is well recognised, for example, between wolf (Canis lupus) and domestic dog (Canis familiaris) (Randi 2008), American bison (Bison bison) and domestic cattle (Bos taurus) (Halbert and Derr 2007), wild and domestic American mink (Neovison vison) (Kidd et al. 2009), European polecat (Mustela putorius) and domestic ferret (Mustela furo) (Davison et al. 1999), wild boar (Sus scrofa) and pig (Sus domesticus) (Scandura et al. 2008) and European wildcat (Felis silvestris) and domestic cat (Felis catus) (e.g., Randi et al. 2001; Pierpaoli et al. 2003). Hybridisation in wildcats is a particularly complex case study, since it involves natural and anthropogenic episodes. Natural hybridisation occurred between the African wildcat (Felis lybica) and other taxa in the wildcat group (Felis spp.) during their evolutionary history (Driscoll et al. 2007; Ottoni et al. 2017; Kitchener et al. 2017). Moreover, F. silvestris and F. lybica were found to hybridise with domestic cats (Driscoll et al. 2007; LeRoux et al. 2015). Domestic cats originally derived from F. lybica in the Near East/North Africa and today the human-mediated dispersal has resulted in a near global distribution (Driscoll et al. 2007; Ottoni et al. 2017).
Once widely distributed, the European wildcat underwent sharp range declines, leading in some cases even to local extinctions by the early twentieth century due to anthropogenic persecution and the loss of suitable habitat (Piechocki 1990; Stahl and Artois 1995; Yamaguchi et al. 2015). Today, the conservation status of the European wildcat is still unfavourable in most European countries (EC 2015), although there is recent evidence of increasing populations and natural recolonisation of the species’ historic range in at least some regions (Steyer et al. 2016; Nussberger et al. 2018). Conservation threats include habitat loss and fragmentation, road mortality, persecution and hybridisation (Klar et al. 2008, 2009; Lozano and Malo 2012; Yamaguchi et al. 2015). However, there is substantial uncertainty about the relative importance of these threats, and in particular, regarding the role of hybridisation.
Previous genetic studies have confirmed the occurrence of hybridisation between wildcats and domestic cats throughout Europe (Randi et al. 2001; Beaumont et al. 2001; Driscoll et al. 2007; Pierpaoli et al. 2003; Lecis et al. 2006; Randi 2008; Oliveira et al. 2008a, b; O’Brien et al. 2009; Hertwig et al. 2009; Eckert et al. 2009; Nussberger et al. 2014b, 2018; Steyer et al. 2018). However, the levels of hybridisation reported varied considerably between studies, even those involving the same regions (e.g., Eckert et al. 2009 or Steyer et al. 2018 vs. Hertwig et al. 2009). This has been suggested to be due to considerable differences in the methods applied to identify hybrid individuals and to measure hybridisation rates, e.g., sampling strategies, size and period, number and type of markers, as well as the statistical approaches used for hybrid identification (Steyer et al. 2018). Recently, polymorphic molecular markers, mainly microsatellites, have been applied for studying admixture and introgression between wildcats and domestic cats (e.g., Randi et al. 2001; Pierpaoli et al. 2003; Eckert et al. 2009). However, the resolution of hybridisation is limited due to the relatively low availability, repeatability between labs and technical capacity to analyse microsatellite markers. High-throughput analyses of single nucleotide polymorphism (SNP) arrays substantially improved the in-depth assessment of hybridisation (e.g., vonHoldt et al. 2013; Goedbloed et al. 2013; Nussberger et al. 2013). SNPs have been shown to be highly accurate and sensitive in identifying hybrid individuals between wildcats and domestic cats, irrespective of origin and available reference database (Oliveira et al. 2015; Steyer et al. 2018; Mattucci et al. 2019).
Here, we analysed wildcat and domestic cat samples from 13 different countries across Europe using a set of ancestry-informative SNP markers (96) to study introgression between wildcats and domestic cats (Nussberger et al. 2013, 2014a). SNP markers were selected to be diagnostic for identifying wildcat, domestic cat, and their hybrids, and included recombinant (autosomal) and non-recombinant markers (mitochondrial and Y-chromosome markers) (Nussberger et al. 2013, 2014a). Autosomal SNPs were analysed using Bayesian statistical tools to identify hybrids and backcrosses. As previous studies have described an asymmetric and sex-specific hybridisation directionality (Nussberger et al. 2018; Oliveira et al. 2018), we analysed paternally inherited (Y-chromosome) and maternally inherited (mitochondrial DNA) SNP markers and sequences. In addition, we genotyped individuals using a set of highly polymorphic microsatellite markers to study the genetic structure of wildcat population.
We present the first large-scale assessment of hybridisation between Felis spp., representing populations across Europe. We aimed to provide a first estimation of overall hybridisation rates in the European wildcat to serve as an initial baseline for future comprehensive assessments. Moreover, we addressed the degree of threat posed by hybridisation to the long-term persistence of wildcats in different regions.
Material and methods
Study area and design
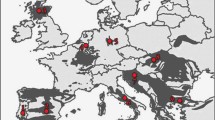
Wildcat and domestic cat samples were collected from 13 countries in Europe between 1999 and 2016. In total, 926 samples were analysed from Southwest and Southern Europe (Portugal n = 69, Spain n = 94, Italy n = 30), Scotland (n = 17), Central Europe (Belgium n = 107, the Netherlands n = 10, Luxembourg n = 48, Germany n = 412, France n = 1, Austria n = 21) and Southeast Europe (Romania n = 59, Bulgaria n = 38, Greece n = 20) (Fig. 1). Samples from Scotland were originally provided as examples of clear morphological hybrids based on pelage, thus reflecting the hybrid swarm that occurs there today (Senn et al. 2018). Therefore, the Scottish samples may be biased more towards hybrids that are closer to domestic cats than wildcats. Samples from Germany were randomly preselected from a larger dataset (Steyer et al. 2018) to avoid local overrepresentation (≤ 4 samples per 10 km2). Sampling was performed opportunistically by collecting samples of carcasses (n = 621), captured cats (n = 65) and domestic cats from pet owners (n = 30), by collecting fresh scat samples (n = 24) and other findings (n = 6), or systematically using hair traps scented with valerian tincture (n = 180) as described by Steyer et al. (2013) (Supplementary Table S1). Sampling material consisted of invasive samples (tissue n = 608, blood n = 88, tooth n = 1) and non-invasive (or minimally invasive) samples (hair n = 202, scat n = 24, saliva n = 3). No animal was harmed or sacrificed for the purposes of this study and all samples were collected in compliance with the respective local and national laws. Genetic samples of captured cats or pets were obtained as byproducts of routine analyses of veterinarians or telemetry studies (Klar et al. 2009; Lammertsma et al. 2011; Streif et al. 2012; Ramos 2014).
Sampling locations and wildcat occurrence in Europe. All genotyped samples (n = 926) are displayed. The colour codes represent populations that were grouped based on microsatellite-based Bayesian clustering. The distribution of the European wildcat (Felis silvestris) and Sardinian wildcat (Felis lybica) in the EU is shown as light grey grid cells (EC 2015)
Laboratory procedures
Extraction of deoxyribonucleic acid (DNA) was performed in separate laboratory rooms for invasive and non-invasive samples. Blood and tissue samples were extracted using the Qiagen Blood and Tissue Kit and the tooth sample was extracted using the Qiagen Investigator Kit following the manufacturer’s instructions. Saliva samples were extracted using the Qiagen QIAamp DNA Blood Mini Kit (Hilden, Germany) following the manufacturer’s instructions and hair samples were extracted using the Qiagen Investigator Kit as described by Steyer et al. (2016). Scat samples were dried at 60 °C for 2 days, followed by subsequent DNA extraction as described by Frantz et al. (2003) and filtering with pre-rinsed Microcon® YM-30 centrifugal filter units (Millipore, Billerica, MA). Negative controls were included in all protocols for detecting potential DNA contamination.
All samples (926) were analysed with a set of 96 SNPs using SNPtype genotyping assays run on Fluidigm 96.96 Dynamic Arrays (Fluidigm, San Francisco, USA), following Nussberger et al. (2014a), to detect admixture between wildcats and domestic cats. The SNP set includes 86 autosomal markers (75 diagnostic markers and 11 markers for individualisation), eight mitochondrial markers and two Y-chromosome markers. All samples were pre-amplified using specific target amplification reactions. The tooth sample and blood and tissue samples were pre-amplified using the manufacturer’s instructions. Pre-amplification of saliva, scat and hair samples was performed according to Nussberger et al. (2014a). No-template controls were included to detect potential contamination on every chip. Raw data were analysed using Fluidigm SNP Genotyping Analysis Software v.3.1.2. The analysis of a subset of 154 samples (17%) was replicated to assess genotyping errors.
A 110 base pair (bp) Felis-specific fragment of the mitochondrial (mt) control region was amplified and sequenced using the primers LF4 (Eckert et al. 2009) and H16498 (Kocher et al. 1989) as described by Steyer et al. (2013). Sequences were processed and aligned in Geneious 6 (https://www.geneious.com, Kearse et al. 2012) using previously published haplotypes (Steyer et al. 2013, 2016) downloaded from GenBank (Accession Numbers: KR076400-KR076428, JX045658-JX045661, KX161418-KX161423).
In total, 905 samples were genotyped with microsatellites to detect population structure. A set of 14 polymorphic microsatellites (Menotti-Raymond et al. 1999) was analysed following the protocol of Steyer et al. (2013). A multiple tube approach with a minimum of three polymerase chain reaction (PCR) replications per sample was applied to measure genotyping errors of potentially low-quality samples (Navidi et al. 1992). Fragment length was analysed using Genemarker 2.2 (SoftGenetics).
Data processing and analyses
The SNP data were filtered for quality in a two-step procedure. Firstly, SNP loci and samples showing more than 90% of missing data were excluded to eliminate markers and samples that generally failed to amplify. Secondly, samples and loci showing more than 30% of missing data were finally removed because an increased rate of failed amplifications (“No call”) has been shown to be related to increased genotyping errors (von Thaden et al. 2017). Genotyping errors were calculated using a customised R-script in the software R 3.2.2. (R Development Core Team 2008) based on the methodology in the software Gimlet (Valiere 2002). Amplification success was calculated for all samples and genotyping errors for replicated samples.
For all replicated SNP and microsatellite genotypes a consensus genotype was built using a customised R-script in the software R (R Development Core Team 2008). An allele was counted if it appeared in at least one out of three replicates, assuming that allelic drop-out occurs more often than false alleles (Kraus et al. 2015; Steyer et al. 2016). Microsatellite genotypes with < 70% amplification success and > 30% allelic drop-out rate were excluded from analysis. Microsatellite genotypes were checked for multiple recorded individuals by using a customised R-script to measure genotype similarities. If available, other criteria, such as mt-haplotype, sex and sampling location or date, were considered. Only one genotype per individual was kept for further analysis.
The software NewHybrids 1.1 (Anderson and Thompson 2002) was used to assess the hybrid status of SNP-typed individuals by analysing genotypes of autosomal SNP markers. The software was configured to estimate posterior probabilities for six different classes: two pure parental groups, F1-hybrids, F2-hybrids and first-generation backcrosses to each parental group. NewHybrids uses a Bayesian framework. We excluded pre-convergence values by discarding (“burn-in”) 100 K iterations, and using the 500 K iterations of each Markov chain (MCMC). Uninformative (i.e. uniform) priors were used as recommended by Steyer et al. (2018). Individuals were considered assigned to one of the six different classes if the assignment value (q(i)) was ≥ 0.85 as described by Steyer et al. (2018).
Two uniparental diagnostic SNPs on the Y-chromosome and eight in the mitochondrial DNA were analysed for paternal and maternal ancestry, respectively. The additionally analysed mitochondrial sequences were aligned and analysed using the software Network 5.0.0.0 to calculate a median joining network (Forster 2015). Mitochondrial SNPs and sequences were aligned further to previously published mitochondrial clades, which covered partly the same segments (ND5, ND6) (Driscoll et al. 2007; Ottoni et al. 2017).
We analysed differentiation among all clusters by DAPC (Discriminant Analysis of Principal Components) in the Adegenet package (Jombart 2008) in R (R Development Core Team 2008) based on all SNP genotypes. We also analysed microsatellite genotypes using the software Structure (Falush et al. 2003) to assess population structure among wildcats only. The Western Central Europe and Central German population were reduced randomly to 35 individuals each to equalize sample sizes between populations. After a “burn-in” of 100 K iterations, inference was based on the values of the remaining 200 K iterations using an admixture model with correlated allele frequencies and no a priori information. Analyses were run for a number of clusters (k) from 1 to 15 in ten independent runs each. The likely number of clusters was determined based on the Evanno method (Evanno et al. 2005) as implemented in the application Structure Harvester (Earl and vonHoldt 2012). Independent runs were joined by using the application Clumpp (Jakobsson and Rosenberg 2007).
Results
Genotyping success and genotyping errors
In total, 926 samples were genotyped with a 96 SNP array. Of those, 32 samples (3%) failed to amplify and 112 samples (12%) had more than 30% of missing data. These samples were excluded from further analysis. Data from 63 autosomal, four mitochondrial and two SRY markers (72%) fulfilled the quality criteria set out above and were thus kept for further analyses. Genotyping errors among the replicated samples (n = 162) showed a rate of 0.7%, representing allelic drop-out and false alleles. Genotyping of 905 samples with microsatellites resulted in a mean amplification success of 85% per sample. The mean allelic drop-out rate for microsatellites genotypes was 6%. We excluded 159 microsatellite genotypes (18%) from further analysis. Repeated detections of the same microsatellite genotype, or if not available of identical SNP genotypes, were assumed to be the same individual and were removed from further analyses (n = 15). In the end, 767 individual genotypes were used for downstream analyses.
Hybridisation analysis
Hybridisation analysis with NewHybrids was run for 767 individuals and 54 autosomal SNP markers, excluding all non-diagnostic markers. In total, 521 individuals were assigned to pure wildcat, 187 to pure domestic cat, four to F1 hybrid, ten to F2 hybrid, 28 to backcross to wildcat, and nine to backcross to domestic cat (Table 1). In eight cases, individuals were not clearly assigned to any of the analysed categories. Hybrids of the first generation (n = 4, F1 hybrids) were significantly less frequent than hybrids of the second generation (n = 47, F2 hybrids and backcrosses). Hybrids were detected in all studied populations with different abundances (Fig. 2). Most hybrids were detected among samples from Scotland. None of the 17 analysed Scottish samples was assigned to wildcat. In other populations the proportion of hybrids among pure wildcats varied between 3 and 21% (Table 2). Besides the Scottish samples, there was no apparent pattern in the geographical distribution of hybrids (Fig. 3). The lowest proportion of hybrids (between 3 and 5%) was found in Western Central Europe, Central Germany and Southeast Europe, especially in the Transylvanian Basin and the Carpathians (Fig. 2, Table 1).
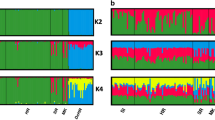
Individual assignments with biparental and uniparental SNPs. All successfully analysed individuals (n = 767) are shown as vertical columns. a Mitochondrial haplotypes were based on a 110 bp sequence of the control region. Classification of haplotypes was according to previous comprehensive studies (Steyer et al. 2016, 2018). b SRY-haplotypes were based on two SRY-SNPs. Classification of the haplotypes followed Nussberger et al. (2014b). c Assignments based on autosomal diagnostic SNPs (n = 54) analysed with the software NewHybrids. Each column represents the individual q(i) value belonging to the respective parental or hybrid cluster. Wildcats (WC) are shown in blue, domestic cats (DC) in red, F1-hybrids (F1) in black, F2-hybrids (F2) in orange, backcrosses to wildcats (BxWC) in light blue and backcrosses to domestic cats (BxDC) in pink and failed amplifications (n/a) in grey. In case of the SRY-SNPs (c) failed amplifications also represent female individuals
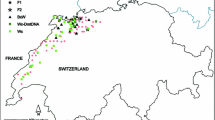
Distribution of wildcats and hybrids in Europe. Assigned categories are based on SNP genotypes (n = 572) analysed with software NewHybrids. Pure wildcats (WC) are shown as blue dots, F1-hybrids (F1) as orange triangles, F2-hybrids (F2) as yellow stars, backcrosses to wildcats (BxWC) as light blue squares and backcrosses to domestic cats (BxDC) as pink squares. Pure domestic cats (n = 187) and samples that could not be assigned to any of the categories (n = 8) are not displayed. The known distribution of wildcats is shown as light grey grid cells (EC 2015)
Variability at uniparental markers (mtDNA and Y-chromosome)
A total of 35 different mitochondrial haplotypes was found among 761 analysed sequences (Fig. 4; GenBank Accession Numbers: KR076400-KR076428, JX045658-JX045661, KX161418-KX161423, MN518925–MN518932). Three main haplogroups were distinguished: a wildcat group (FS-A), a domestic cat group (FS-B) and a group shared between both (FS-C). The highest frequency of the haplogroup FS-C was found in Southeast and Central Europe (Supplementary Fig. S1). Within the groups, private haplotypes were found for wildcats and domestic cats with very few cytonuclear discordances (n = 18) (Fig. 4). Of identified hybrids, 88% showed wildcat haplotypes (Fig. 2). Among the Scottish samples 15 out of 17 individuals (88%) carried the same wildcat haplotype (Supplementary Fig. S2).
Network of mitochondrial haplotypes with corresponding hybrid category. The network was calculated with a 110 bp fragment of the control region. Black dots between haplotypes display the number of mutation steps (added by 1). Each number per haplotype is concordant with the name provided in GenBank (SGN-HP-FS03 to -FS60), except for haplotypes which have been detected for the first time (SGN-FS63 to -74). The pie charts per haplotype indicate the assigned hybrid category based on autosomal SNP genotypes (n = 759) analysed with the software NewHybrids. The size of the pie charts corresponds to the number of detections
Based on the mitochondrial SNPs, clades I and IV were identified corresponding to the classification of Ottoni et al. (2017) (Supplementary Fig. S3). Clade I was exclusively found in wildcats and clade IV appeared in wildcats and domestic cats.
The two SRY-SNPs showed only two different combinations (Y/Y or X/X) classified as domestic cat or wildcat type according to Nussberger et al. (2013). Among all analysed samples Y-chromosomes of wildcat type were found in 417 males and of domestic cat type in 133 males. Within these there was a high concordance with assignments based on autosomal SNP markers (Fig. 2). In case of the SRY SNPs 15 individuals (3%) showed discordant results; four domestic cats carried a Y-chromosome of wildcat ancestry, whilst 11 wildcats carried one of domestic cat ancestry. Seventeen detected male hybrids carried a Y-chromosome of wildcat ancestry and 15 of domestic cat ancestry. Among the Scottish samples 9 out of 11 males (81%) carried a domestic cat Y-chromosome (Fig. 2).
Population genetic structure
Results of a clustering analysis in DAPC, using all SNP genotypes, were concordant with those in NewHybrids. Wildcats and domestic cats plotted into two distinct groups and hybrids appeared admixed (Supplementary Fig. S4). SNP markers were highly discriminating between wildcats and domestic cats but indicated low informative value (eigenvalue) concerning more fine-scale population structures.
The polymorphic set of 14 microsatellite markers revealed distinct population structures among wildcat populations in Structure. The most likely number of genetic clusters was k = 2, revealing a clear distinction of wildcats in Central Germany from other populations (Supplementary Fig. S5). High likelihood was also achieved for k = 3, k = 4 and k = 5. In total, five genetic clusters were differentiated and grouped into populations throughout the study area: Central Germany, Iberian Peninsula, Western Central Europe (Western Germany, Belgium, Netherlands, Luxembourg, France), Eastern Alpine (Austria, Northern Italy) and Southeast Europe (Romania, Bulgaria, Greece) (Fig. 1). Individuals from Romania, Bulgaria and Greece did not reveal population substructure. The individuals from central Italy (n = 8) appeared admixed between clusters and were grouped as a separate population consistent with previous studies (Mattucci et al. 2013). Individuals from Scotland were also grouped separately because of the lack of samples assigned as wildcats. In total, samples were grouped into seven populations throughout the study area (Fig. 1).
Discussion
The aim of this study was to assess hybridisation levels between the European wildcat and the domestic cat throughout Europe and to compare findings between different areas of its distribution. The 51 hybrids were found among 521 wildcat individuals throughout the wildcat’s distribution in Europe (Fig. 3). Levels of hybridisation varied considerably in frequency and distribution in the populations considered (Table 1). They were low to moderate in Central, Southeast and Southwest Europe. In contrast, all samples from Scotland were identified as backcross hybrids, supporting findings of previous studies that the genetic integrity of the wildcat population in Scotland is seriously compromised (Kitchener et al. 2005; Kitchener and Daniels 2008; Kilshaw et al. 2016; Senn et al. 2018). However, since the Scottish samples were explicitly morphological hybrids, this may have biased the result. On the other hand, the lack of cats with typical wildcat appearance probably reflects the hybrid swarm status of this population.
In most populations, maternal and paternal haplotypes were both coincident with the autosomal DNA result, which suggests that there have been no intense hybridisation episodes in the past. Hence, our SNP analyses confirm previous findings based on microsatellites that the genetic integrity of the European wildcat has persisted in most regions to date (e.g., Randi et al. 2001; Pierpaoli et al. 2003; Mattucci et al. 2016). Most Scottish samples carried a mitochondrial haplotype of wildcat type and Y-chromosomes of domestic cat type, which may reflect a sex-biased directionality of gene flow. However, this may also result from the limited sample size or reflect a potential bias in these Scottish samples towards the advanced level of introgression within this population (Senn et al. 2018).
Irrespective of these findings, it remains unclear to which degree both forms are affected by historic gene flow because available current hybridisation assessments are restricted to measuring contemporary differentiation between wildcats and domestic cats. Palaeogenetic or genomic studies investigating proportions of admixture in wildcats need to be developed. Mattucci et al. (2019) recently developed an approach to detect genomic traces originating from hybridisation events that occurred from 6 to 22 generations in the past.
While we found hybrids throughout all studied wildcat populations, we also assessed low to moderate levels of hybridisation in most regions. Hybridisation levels, assessed with SNP data, were generally similar to previously reported results based on microsatellite data (summarised in Table 2) The lowest proportion of hybrids (3–5%) was detected in Central Europe, which is supported by previous studies (Pierpaoli et al. 2003; Hertwig et al. 2009; Eckert et al. 2009; Steyer et al. 2018), excepting for a high hybridisation rate of 43% for western Germany, described by Hertwig et al. (2009). This finding was not supported here and in previous studies covering the same study areas (Pierpaoli et al. 2003; Eckert et al. 2009; Steyer et al. 2016). Steyer et al. (2018) suggested that the discrepancy derives from methodological differences related to the problem of reference population assignment when relying on microsatellite data. In Southeast Europe, we found similarly low hybridisation levels (5%), which are the first genetically confirmed results covering several regions in this area (Table 2). Findings of moderate hybridisation rates in the Iberian Peninsula were in line with previous findings that were based on microsatellite analyses (Table 2; Pierpaoli et al. 2003; Oliveira et al. 2008a, b). In the Eastern Alpine region, we detected one F1 and two backcross hybrids among 23 individuals, which are, to our knowledge, the first genetically confirmed hybrids in this area. In Scotland, we found exclusively hybrids of different classes, indicating that hybridisation has been occurring for several generations and crossbreeding between different hybrid classes (Kitchener et al. 2005; Kilshaw et al. 2016; Mattucci et al. 2016; Senn et al. 2018).
Most previous studies addressing recent hybridisation between wildcats and domestic cats were based on regional sampling and/or relied on limited sets of microsatellite markers (usually 14 or less). Broad-scale comparisons of hybridisation rates had low informative value due to methodological constraints, namely to the use of different markers, statistical approaches and thresholds. The application of SNPs provides several advantages, namely that they are abundant and broadly distributed in genomes and compatible with high-throughput approaches (reviewed by Garvin et al. 2010). The applied SNP-set has been previously shown to be statistically powerful in differentiating wildcats, domestic cats and F1, F2 and first-generation backcrosses (correct assignments of 99.6% (Nussberger et al. 2013); correct assignments of F1 hybrids 100%, second generation hybrids 96–98% (Steyer et al. 2018)). The microsatellite set analysed in this study has been previously shown to fail in differentiating among hybrid classes (Steyer et al. 2018) and was thus not considered in the assessment of hybridisation. However, obstacles in comparing hybridisation rates are not completely overcome, including diverse use of reference populations, differences in sampling techniques and intensity throughout the species distribution (Steyer et al. 2018). Future research needs to address a harmonisation of sampling approaches for European wildcats for conducting more comprehensive assessments.
While admixture between European wildcats and domestic cats was widespread throughout the species’ range, we found low or moderate levels of recent hybridisation in most regions. If only F1 hybrids are considered, the overall proportion of hybrids among studied samples was less than 1%, since most hybrids were F2 and backcrosses. However, the very low frequency of observed F1s should be taken with caution, since this can reflect a sample bias (animals too similar to domestic cats may be not sampled). Overall, our findings generally suggest that the genetic integrity of European wildcats persists in the long term despite hybridisation events. Conversely, we confirmed regionally elevated introgression rates, as in Scotland: all analysed samples were identified as backcrossed hybrids. In this population, a high proportion of F2 hybrids (29%) was also detected, although this is likely to be due to a sampling bias, since F2 hybrids (F1 × F1) are expected to occur in rare incidences only. The use of strict hybrid categories might have caused the misassignments of individuals that are descended from repeated crossbreeding between different hybrid generations (see also Senn et al. 2018). These findings confirm that hybridisation has been continuing for several generations.
Increased hybridisation rates, leading to considerable introgression, have significant consequences for the conservation of wildcat populations (Yamaguchi et al. 2015). Considering the emblematic case of the Scottish European wildcat population, it is important to understand the factors that affect the frequency of hybridisation. Domestic cats have a worldwide distribution and the number of pet cats exceeds 65 million in European countries with wildcat presence (EPFI 2017). The total number of domestic cats is estimated to be higher by several orders of magnitude than that of European wildcats. Differences in population sizes between hybridising taxa, as seen for wildcat and domestic cat in Europe, may increase the likelihood of extinction of the smaller population (Rhymer and Simberloff 1996). The actual number of free-ranging domestic cats is unknown because some pet cats are kept indoors and solid estimates of feral domestic cats are lacking. The degree of dependence of domestic cats on humans is expected to be highly variable and it is generally difficult to assess data on feral cats.
Considering the hybridisation levels for mainland Europe and the level of anthropogenic disturbance in the area, we think that ecological and/or ethological factors, limiting panmixia between both forms, must exist in most populations. Previous studies have reported that hybrids occurred more frequently at the periphery of the ranges of wildcats (Randi et al. 2001), wolves (Randi 2008) or golden jackals (Canis aureus; Galov et al. 2015). The peripheral parts of populations are expected to have lower population densities than core areas, which may affect ethological factors affecting mate choice, also known as the Allee effect (Allee 1931). Previous studies have shown that recent range expansions of wildcat populations have led to increased hybridisation rates in Switzerland and France (Nussberger et al. 2014b, 2018). As male cats generally disperse farther than females (Sunquist and Sunquist 2002), an asymmetric distribution of maternal or paternal markers can reflect population-dynamic processes. In addition, male wildcats have been shown to tolerate low quality habitat better compared to females, which may also affect directionality of hybridisation (Oliveira et al. 2018). In the Jura region of eastern Switzerland and western France, an increased rate of introgression of domestic cat mitochondrial haplotypes into wildcats was found and explained by a sex-biased dispersal of male wildcats entering domestic cat ranges (Nussberger et al. 2014b, 2018). Among the Scottish samples, we found an opposite pattern of hybrids, which mostly carry a domestic-cat-type Y-chromosome but a wildcat-type mitochondrial haplotype, which may suggest an opposite sex-biased directionality of gene flow. This may be explained by prevalent gene flow between hybrids and male domestic cats and female wildcats, but this finding needs to be confirmed on a larger sampling set. Interestingly, hybrids have been observed to occupy the same habitat as wildcats and partly the same as domestic cats (Germain et al. 2008; Kilshaw et al. 2016). Hence, hybrids may play a role as vectors for gene flow between both populations, accelerating admixture between both species.
Encounters between wildcats and domestic cats are expected to be a result of individual movements, which are affected by habitat and population status (Gil-Sánchez et al. 2015). Movement data on both wildcats and domestic cats have shown low spatio-temporal overlap on a local scale (Germain et al. 2008). In Central Europe, wildcats have been observed to prefer habitats with a proximity to forests and a critical distance of several hundred meters from villages, single houses and roads to avoid human disturbance (Klar et al. 2008). In contrast, domestic cats prefer habitats in proximity to human settlements, because of access to provided resources (Biró et al. 2004; Ferreira et al. 2011), while feral domestic cats may be less dependent on supplied food resources. Consequently, encounters between wildcats and domestic cats may occur frequently at the edges or outside preferred wildcat habitat. In addition, encounters may take place through exploratory movements outside their home ranges, particularly during the mating season (Germain et al. 2008). Domestic cats, for instance, have been detected within protected areas at a considerable distance from human settlements (e.g., Sarmento et al. 2009; Zwijacz-Kozica et al. 2017). Wildcats and domestic cats have also been shown to occur at the same locations, at least occasionally (Nussberger et al. 2014b; Kilshaw et al. 2016; Steyer et al. 2016; Beutel et al. 2017). We assume that habitat fragmentation may enhance the chance of encounters between wildcats and domestic cats because of the higher proportion of land being subject to an edge effect. The high hybridisation rate in Hungary has been explained by the occurrence of wildcats in highly fragmented areas of forest patches, agriculture and human settlements (Pierpaoli et al. 2003; Lecis et al. 2006; Randi 2008). In Central Europe, we found a low hybridisation rate (Table 2) despite a relatively high level of landscape fragmentation. This result may be explained by the persistence of considerable amounts of broad-leaf and mixed forest habitats, particularly in the moderately populated low mountain regions (Steyer et al. 2018).
Hybridisation between the European wildcat and the early forms of domestic cat may have been occurring in Europe since the spread of Neolithic farming (Ottoni et al. 2017). There is evidence for prehistoric gene flow between members of the Felis silvestris/lybica species complex, suggesting a complicated phylogenetic relationship (Driscoll et al. 2007; Ottoni et al. 2017). In our study, we analysed non-recombinant paternal markers to assess current distribution and frequency of uniparental lineages in European wildcats and domestic cats. Interestingly, the assignment of mitochondrial haplotypes revealed that wildcats and domestic cats do not appear as completely distinct maternal clades. Besides a wildcat clade (FS-A) and a domestic cat clade (FS-B), there is a shared one between both taxa (FS-C; Fig. 4). Similar findings have been described in previous studies (Driscoll et al. 2007; Eckert et al. 2009; Steyer et al. 2016, 2018).
In our study, the individual haplotypes within the clade FS-C appeared exclusively for domestic cats or wildcats. Hence, a derivation from recently occurring hybridisation is highly unlikely. The two wildcat haplotypes within the clade FS-C (FS22 and FS23) occurred commonly in several studied wildcat populations showing highest frequencies in Southeast Europe and Central Germany. Baca et al. (2018) suggested that ancient hybridisation may have led to introgression from early arriving domestic cats to wildcats. Ottoni et al. (2017) found evidence for ancient gene flow taking place between F. silvestris and F. lybica, the ancestor of domestic cats. They suggest that the range of F. lybica extended temporarily beyond the formerly existing Bosphorus land bridge between Europe and Asia due to climatic fluctuations during Late Pleistocene (Ottoni et al. 2017). During the last glacial period populations of the European wildcat drastically declined and persisted in several small refugia in southern Europe (Sommer and Benecke 2006). When domestic cats firstly arrived in Southeast Europe from around 6000 years ago (Ottoni et al. 2017), the postglacial range expansion of the European wildcat already included large parts of Central Europe (Sommer and Benecke 2006). The clear prevalence of haplotypes FS22 and FS23 in wildcat populations in Southeast and Central Europe today may support an introgression event that occurred before the postglacial expansion of the European wildcat. However, a derivation from incomplete lineage sorting, as suggested by Eckert et al. (2009), may lead to similar patterns and thus serves as another explanation. Considering the complex phylogenetic relations between members of the Felis silvestris/lybica species complex, it is highly recommended to include recombinant nuclear markers when identifying species or hybrids and, if available, also to use diagnostic morphological characters.
Conclusions and implications for conservation
Using a panel of discriminative SNP markers, we confirmed that hybridisation between wildcats and their domestic congeners is a widespread phenomenon throughout the species’ range. However, the overall level of hybridization is moderate, suggesting that long-term coexistence with the domestic cat may allow the persistence of the genetic integrity of wildcat populations, even in anthropogenically disturbed landscapes with high abundances of domestic cats. Nevertheless, frequent hybridisation with the domestic cat may regionally threaten the genetic integrity of the European wildcat, as documented by the example of the wildcat in Scotland and potentially leading even to the genetic extinction of local populations.
Our findings highlight the need for regionally adapted conservation management for wildcats, which accounts for the geographically varying importance of hybridisation as a threat for the long-term integrity of the species. Regular monitoring of wildcat populations and hybridisation rates is highly recommended. We strongly urge the application of harmonised nuclear marker panels throughout Europe to achieve supra-regional comparisons of hybridisation rates and degrees of introgression in local wildcats. Only the collection of further large-scale data allows a deeper insight into the reasons and mechanisms of regionally accelerated hybridisation rates in different regions and particularly at the edges of the species’ distribution that may be more susceptible to hybridisation. To understand the dynamics and consequences of hybridisation between wildcats and domestic cats, we highlight the need for interdisciplinary research involving palaeobiology, landscape ecology, ethology and genomics.
Data accessibility
Sampling locations, all genetic raw data, and results from the software analyses as reported in this study, including mitochondrial sequences, genotypes from microsatellites, and SNPs, are available in supplementary information files. DNA sequence data used for haplotype network have been submitted to GenBank and have accession numbers KR076400-KR076428, JX045658-JX045661, KX161418-KX161423, and MN518925-MN518932. Customized R-Script are available from the corresponding authors on reasonable request.
References
Abbott RJ, Barton NH, Good JM (2016) Genomics of hybridization and its evolutionary consequences. Mol Ecol 25:2325–2332. https://doi.org/10.1111/mec.13685
Allee WC (1931) Animal aggregations. A study in General Sociology. University of Chicago Press, Chicago (Illinois)
Allendorf FW, Leary RF, Spruell P, Wenburg JK (2001) The problems with hybrids: setting conservation guidelines. Trends Ecol Evol 16:613–622
Anderson EC, Thompson EA (2002) A model-based method for identifying species hybrids using multilocus genetic data. Genetics 160:1217–1229
Baca M, Popović D, Panagiotopoulou H, Marciszak A, Krajcarz M, Krajcarz MT, Makowiecki D, Węgleński P, Nadachowski A (2018) Human-mediated dispersal of cats in the Neolithic Central Europe. Heredity. https://doi.org/10.1101/259143
Beaumont M, Barratt EM, Gottelli D, Kitchener AC, Daniels MJ, Pritchard JK, Bruford MW (2001) Genetic diversity and introgression in the Scottish wildcat. Mol Ecol 10:319–336
Beutel T, Reineking B, Tiesmeyer A, Nowak C, Heurich M (2017) Spatial patterns of co-occurrence of the European wildcat Felis silvestris silvestris and domestic cats Felis silvestris catus in the Bavarian Forest National Park. Wildlife Biol. https://doi.org/10.2981/wlb.00284
Bidlack AL, Reed SE, Palsbøll PJ, Getz WM (2007) Characterization of a western North American carnivore community using PCR–RFLP of cytochrome b obtained from fecal samples. Conserv Genet 8:1511–1513
Biró Z, Szemethy L, Heltai M (2004) Home range sizes of wildcats (Felis silvestris) and feral domestic cats (Felis silvestris f. catus) in a hilly region of Hungary. Mamm Biol 69:302–310
Bramanti B, Thomas MG, Haak W, Unterlaender M, Jores P, Tambets K, Antanaitis-Jacobs I, Haidle MN, Jankauskas R, Kind C-J, Lueth F, Terberger T, Hiller J, Matsumura S, Forster P, Burger J (2009) Genetic discontinuity between local hunter-gatherers and Central Europe's first farmers. Science 326:137–140
Davison A, Birks J, Griffiths H, Kitchener A, Biggins D, Butlin R (1999) Hybridization and the phylogenetic relationship between polecats and domestic ferrets in Britain. Biol Conserv 87:155–161
Driscoll CA, Menotti-Raymond M, Roca AL, Hupe K, Johnson WE, Geffen E, Harley EH, Delibes M, Pontier D, Kitchener AC, Yamaguchi N, O’Brien SJ, Macdonald DW (2007) The near Eastern origin of cat domestication. Science 317:519–523
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361. https://doi.org/10.1007/s12686-011-9548-7
Eckert I, Suchentrunk F, Markov G, Hartl GB (2009) Genetic diversity and integrity of German wildcat (Felis silvestris) populations as revealed by microsatellites, allozymes, and mitochondrial DNA sequences. Mamm Biol 75:160–174
European Commission (2015) Reporting from EU Member States under Article 17 of the Habitats Directive to the European Commission. https://www.eea.europa.eu/data-and-maps/data/article-17-database-habitats-directive-92-43-eec-1#tab-metadata. Accessed 17 July 2018
European Pet Food Industry (2017) Facts and figures 2017. https://www.fediaf.org/who-we-are/european-statistics.html. Accessed 17 July 2018
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 4:2611–2620
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Ferreira JP, Leitão I, Santos-Reis M, Revilla E (2011) Human-related factors regulate the spatial ecology of domestic cats in sensitive areas for conservation. PLoS ONE. https://doi.org/10.1371/journal.pone.0025970
Forster M (2015) Network 5.0.0.0. Fluxus Technology. fluxus-engineering.com
Frantz AC, Pope LC, Carpenter PJ, Roper TJ, Wilson GJ, Delahay RJ, Burke T (2003) Reliable microsatellite genotyping of the Eurasian badger (Meles meles) using faecal DNA. Mol Ecol 12:1649–1661
Futuyma DJ (2005) Evolution. Sinauer Associates, Sunderland
Galov A, Fabbri E, Caniglia R, Arbanasić H, Lapalombella S, Florijančić T, Bošković I, Galaverni M, Randi E (2015) First evidence of hybridization between golden jackal (Canis aureus) and domestic dog (Canis familiaris) as revealed by genetic markers. R Soc Open Sci. https://doi.org/10.1098/rsos.150450
Garvin MR, Saitoh K, Gharrett AJ (2010) Application of single nucleotide polymorphisms to non-model species: a technical review. Mol Ecol Resour 10:915–934. https://doi.org/10.1111/j.1755-0998.2010.02891.x
Germain E, Benhamou S, Poulle ML (2008) Spatio-temporal sharing between the European wildcat, the domestic cat and their hybrids. J Zool 276:195–203
Gil-Sánchez JM, Jaramillo J, Barea-Azcón JM (2015) Strong spatial segregation between wildcats and domestic cats may explain low hybridization rates on the Iberian Peninsula. Zoology 118:377–385. https://doi.org/10.1016/j.zool.2015.08.001
Goedbloed DJ, Megens HJ, van Hooft P, Herrero-Medrano JM, Lutz W, Alexandri P, Crooijmans RPMA, Groenen MA, Van Wieren SE, Ydenberg RC, Prins HHT (2013) Genome-wide single nucleotide polymorphism analysis reveals recent genetic introgression from domestic pigs into Northwest European wild boar populations. Mol Ecol 22:856–866. https://doi.org/10.1111/j.1365-294X.2012.05670.x
Halbert ND, Derr JN (2007) A comprehensive evaluation of cattle introgression into US federal bison herds. J Hered 98:1–12
Hertwig ST, Schweizer M, Stepanow S, Jungnickel A, Bohle U-R, Fischer MS (2009) Regionally high rates of hybridization and introgression in German wildcat populations (Felis silvestris, Carnivora, Felidae). J Zool Syst Evol Res 47:283–297. https://doi.org/10.1111/j.1439-0469.2009.00536.x
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806. https://doi.org/10.1093/bioinformatics/btm233
Jombart T (2008) adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. https://doi.org/10.1093/bioinformatics/bts199
Kidd AG, Bowman J, Lesbarrères D, Schulte-Hostedde AI (2009) Hybridization between escaped domestic and wild American mink (Neovison vison). Mol Ecol 18:1175–1186. https://doi.org/10.1111/j.1365-294X.2009.04100
Kilshaw K, Montgomery RA, Campbell RD, Hetherington DA, Johnson PJ, Kitchener AC, Macdonald DW, Millspaugh JJ (2016) Mapping the spatial configuration of hybridization risk for an endangered population of the European wildcat (Felis silvestris silvestris) in Scotland. Mamm Res 61:1–11. https://doi.org/10.1007/s13364-015-0253-x
Kitchener AC, Daniels MJ (2008) Wildcat Felis silvestris. In: Harris S, Yalden DW (eds) Mammals of the British Isles: Handbook, 4th edn. The Mammal Society, Southampton, pp 397–406
Kitchener AC, Yamaguchi N, Ward JM, Macdonald DW (2005) A diagnosis for the Scottish wildcat (Felis silvestris): a tool for conservation action for a critically-endangered felid. Anim Conserv 8:223–237. https://doi.org/10.1017/S1367943005002301
Kitchener AC, Breitenmoser-Würsten C, Eizirik E, Gentry A, Werdelin L, Wilting A, Yamaguchi N, Abramov AV, Christiansen P, Driscoll C, Duckworth JW, Johnson W, Luo, SJ, Meijaard E, O’Donoghue P, Sanderson J, Seymour K, Bruford M, Groves C, Hoffmann M, Nowell K., Timmons Z, Tobe S (2017) A revised taxonomy of the Felidae. The final report of the Cat Classification Task Force of the IUCN/SSC Cat Specialist Group. Cat News Special Issue 11
Klar N, Fernández N, Kramer-Schadt S, Herrmann M, Trinzen M, Büttner I, Niemitz C (2008) Habitat selection models for European wildcat conservation. Biol Conserv 141:308–319
Klar N, Herrmann M, Kramer-Schadt S (2009) Effects and mitigation of road impacts on individual movement behavior of wildcats. J Wildl Manage 73:631–638
Kocher TD, Thomas WK, Meyer A, Edwards SV, Pääbo S, Villablanca FX, Wilson AC (1989) Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers. Proc Natl Acad Sci USA 86:6196–6200
Kraus RHS, vonHoldt B, Cocchiararo B, Harms V, Bayerl H, Kuhn R, Forster DW, Fickel J, Roos C, Nowak C (2015) A single-nucleotide polymorphism-based approach for rapid and cost-effective genetic wolf monitoring in Europe based on noninvasively collected samples. Mol Ecol Resour 15:295–305. https://doi.org/10.1111/1755-0998.12307
Lammertsma DR, Janssen R, van der Hout J, Jansman HAH (2011) Huiskatten in natuurgebieden; Kan TNR hybridisatie met de Wilde kat voorkomen? Alterra, Wageningen, Netherlands. Alterra-rapport 2263
Le Roux JJ, Foxcroft LC, Herbst M, MacFadyen S (2015) Genetic analysis shows low levels of hybridization between African wildcats (Felis silvestris lybica) and domestic cats (F. s. catus) in South Africa. Ecol Evol 5:288–299. https://doi.org/10.1002/ece3.1275
Lecis R, Pierpaoli M, Biró ZS, Szemethy L, Ragni B, Vercillo F, Randi E (2006) Bayesian analyses of admixture in wild and domestic cats (Felis silvestris) using linked microsatellite loci. Mol Ecol 15:119–131
Lozano J, Malo AF (2012) Conservation of the European wildcat (Felis silvestris) in Mediterranean environments: a reassessment of current threats. In: Williams GS (ed) Mediterranean ecosystems: dynamics, management and conservation. Nova Science Publisher's Inc., Hauppauge, pp 2–31
Mallet J (2008) Hybridization, ecological races and the nature of species: Empirical evidence for the ease of speciation. Phil Trans R Soc Lond Ser B 363:2971–2986. https://doi.org/10.1098/rstb.2008.0081
Mattucci F, Oliveira R, Bizzarri L, Vercillo F, Anile S, Ragni B, Lapini L, Sforzi A, Alves PC, Lyons LA, Randi E (2013) Genetic structure of wildcat (Felis silvestris) populations in Italy. Ecol Evol 3:2443–2458. https://doi.org/10.1002/ece3.569
Mattucci F, Oliveira R, Lyons LA, Alves PC, Randi E (2016) European wildcat populations are subdivided into five main biogeographic groups: consequences of Pleistocene climate changes or recent anthropogenic fragmentation? Ecol Evol 6:3–22. https://doi.org/10.1002/ece3.1815
Mattucci F, Galaverni M, Lyons LA, Alves PC, Randi E, Velli E, Pagani L, Caniglia R (2019) Genomic approaches to identify hybrids and estimate admixture times in European wildcat populations. Sci Rep 9:11612. https://doi.org/10.1038/s41598-019-48002-w
Menotti-Raymond M, David VA, Lyons LA, Schäffer AA, Tomlin JF, Hutton MK, O’Brien SJ (1999) A genetic linkage map of microsatellites in the domestic cat (Felis catus). Genomics 57:9–23
Mooney HA, Cleland EE (2001) The evolutionary impact of invasive species. Proc Natl Acad Sci USA 98:5446–5451
Navidi W, Arnheim N, Waterman MS (1992) A multiple-tubes approach for accurate genotyping of very small DNA samples by using PCR: statistical considerations. Am J Hum Genet 50:347–359
Nussberger B, Greminger MP, Grossen C, Keller LF, Wandeler P (2013) Development of SNP markers identifying European wildcats, domestic cats, and their admixed progeny. Mol Ecol Resour 13:447–460. https://doi.org/10.1111/1755-0998.12075
Nussberger B, Wandeler P, Camenisch G (2014a) A SNP chip to detect introgression in wildcats allows accurate genotyping of single hairs. Eur J Wildl Res 60:405–410. https://doi.org/10.1007/s10344-014-0806-3
Nussberger B, Wandeler P, Weber D, Keller LF (2014b) Monitoring introgression in European wildcats in the Swiss Jura. Conserv Genet 15:1219–1230. https://doi.org/10.1007/s10592-014-0613-0
Nussberger B, Currat M, Quilodran CS, Ponta N, Keller LF (2018) Range expansion as an explanation for introgression in European wildcats. Biol Conserv 218:49–56. https://doi.org/10.1016/j.biocon.2017.12.009
O’Brien J, Devillard S, Say L, Vanthomme H, Léger F, Ruette S, Pontier D (2009) Preserving genetic integrity in a hybridising world: are European Wildcats (Felis silvestris silvestris) in eastern France distinct from sympatric feral domestic cats? Biodivers Conserv 18:2351–2360. https://doi.org/10.1007/s10531-009-9592-8
Oliveira R, Godinho R, Randi E, Alves PC (2008a) Hybridization versus conservation: are domestic cats threatening the genetic integrity of wildcats (Felis silvestris silvestris) in Iberian Peninsula? Phil Trans R Soc Lond Ser B 363:2953–2961. https://doi.org/10.1098/rstb.2008.0052
Oliveira R, Godinho R, Randi E, Ferrand N, Alves PC (2008b) Molecular analysis of hybridisation between wild and domestic cats (Felis silvestris) in Portugal: implications for conservation. Conserv Genet 9:1–11. https://doi.org/10.1007/s10592-007-9297-z
Oliveira R, Randi E, Mattucci F, Kurushima JD, Lyons LA, Alves PC (2015) Toward a genome-wide approach for detecting hybrids: informative SNPs to detect introgression between domestic cats and European wildcats (Felis silvestris). Heredity 115:195–205. https://doi.org/10.1038/hdy.2015.25
Oliveira T, Urra F, López-Martín JM, Ballesteros-Duperón E, Barea-Azcón JM, Moléon M, Gil-Sánchez JM, Alves PC, Díaz-Ruíz F, Ferreras P, Monterroso P (2018) Females know better: sex-biased habitat selection by the European wildcat. Ecol Evol 8:9464–9477. https://doi.org/10.1002/ece3.4442
Ottoni C, van Neer W, de Cupere B et al (2017) The palaeogenetics of cat dispersal in the ancient world. Nat Ecol Evol. https://doi.org/10.1038/s41559-017-0139
Paxinos E, McIntosh C, Ralls K, Fleischer R (1997) A noninvasive method for distinguishing among canid species: amplification and enzyme restriction of DNA from dung. Mol Ecol 6:483–486. https://doi.org/10.1046/j.1365-294X.1997.00206.x
Piechocki R (1990) Die Wildkatze: Felis silvestris, 1. Aufl. Die neue Brehm-Bücherei, vol 189. Ziemsen, Wittenberg Lutherstadt.
Pierpaoli M, Biró ZS, Herrmann M, Hupe K, Fernandes M, Ragni B, Szemethy L, Randi E (2003) Genetic distinction of wildcat (Felis silvestris) populations in Europe, and hybridization with domestic cats in Hungary. Mol Ecol 12:2585–2598. https://doi.org/10.1046/j.1365-294X.2003.01939.x
R Development Core Team (2008) R: a language and environment for statistical computing. Vienna, Austria. R Foundation for Statistical Computing. https://www.R-project.org
Ramos L (2014) Assessing hybridization between wildcat and domestic cat: the particular case of Iberian Peninsula and some insights into North Africa. Master’s thesis. University of Porto.
Randi E (2008) Detecting hybridization between wild species and their domesticated relatives. Mol Ecol 17:285–293. https://doi.org/10.1111/j.1365-294X.2007.03417.x
Randi E, Pierpaoli M, Beaumont M, Ragni B, Sforzi A (2001) Genetic identification of wild and domestic cats (Felis silvestris) and their hybrids using Bayesian clustering methods. Mol Biol Evol 18:1679–1693
Rhymer JM, Simberloff D (1996) Extinction by hybridization and introgression. Annu Rev Ecol Evol Syst 27:83–109
Sakai AK, Allendorf FW, Holt JS, Lodge DM, Molofsky J, With KA, Baughman S, Cabin RJ, Cohen JE, Ellstrand NC, McCauley DE, O'Neil P, Parker IM, Thompson JN, Weller SG (2001) The population biology of invasive species. Annu Rev Ecol Evol Syst 32:305–332. https://doi.org/10.1146/annurev.ecolsys.32.081501.114037
Sarmento P, Cruz J, Eira C, Fonseca C (2009) Spatial colonization by feral domestic cats Felis catus of former wildcat Felis silvestris silvestris home ranges. Acta Theriol (Warsz) 54:31–38
Say L, Devillard S, Léger F, Pontier D, Ruette S (2012) Distribution and spatial genetic structure of European wildcat in France. Anim Conserv 15:18–27. https://doi.org/10.1111/j.1469-1795.2011.00478.x
Scandura M, Iacolina L, Crestanello B, Pecchioli E, Di Benedetto MF, Russo V, Davoli R, Apollonio M, Bertorelle G (2008) Ancient vs. recent processes as factors shaping the genetic variation of the European wild boar: are the effects of the last glaciation still detectable? Mol Ecol 17:1745–1762. https://doi.org/10.1111/j.1365-294X.2008.03703.x
Seehausen O, Takimoto G, Roy D, Jokela J (2008) Speciation reversal and biodiversity dynamics with hybridization in changing environments. Mol Ecol 17:30–44. https://doi.org/10.1111/j.1365-294X.2007.03529.x
Senn H, Ghazali M, Kaden J, Barclay D, Harrower B, Campbell RD, Macdonald DW, Kitchener AC (2018) Distinguishing the victim from the threat: SNP-based methods reveal the extent of introgressive hybridisation between wildcats and domestic cats in Scotland and inform future in-situ and ex-situ management options for species restoration. Evol Appl. https://doi.org/10.1111/eva.12720
Simberloff D, Martin J-L, Genovesi P, Maris V, Wardle DA, Aronson J, Courchamp F, Galil B, García-Berthou E, Pascal M, Pyšek P, Sousa R, Tabacchi E, Vilà M (2013) Impacts of biological invasions: what's what and the way forward. Trends Ecol Evol 28:58–66. https://doi.org/10.1016/j.tree.2012.07.013
Sommer RS, Benecke N (2006) Late Pleistocene and Holocene development of the felid fauna (Felidae) of Europe: a review. J Zool 269:7–19. https://doi.org/10.1111/j.1469-7998.2005.00040.x
Stahl P, Artois M (1995) Status and conservation of the wildcat (Felis silvestris) in Europe and around the Mediterranean rim. Nature and environment, vol 69. Council of Europe, Strasbourg.
Steyer K, Simon O, Kraus RHS, Haase P, Nowak C (2013) Hair trapping with valerian-treated lure sticks as a tool for genetic wildcat monitoring in low-density habitats. Eur J Wildl Res 59:39–46. https://doi.org/10.1007/s10344-012-0644-0
Steyer K, Kraus RHS, Mölich T et al (2016) Large-scale genetic census of an elusive carnivore, the European wildcat (Felis s. silvestris). Conserv Genet 17:1183–1199. https://doi.org/10.1007/s10592-016-0853-2
Steyer K, Tiesmeyer A, Muñoz-Fuentes V, Nowak C (2018) Low rates of hybridization between European wildcats and domestic cats in a human-dominated landscape. Ecol Evol 8:2290–2304. https://doi.org/10.1002/ece3.3650
Streif S, Kraft S, Veith S, Kohnen A, Suchant R (2012) Monitoring and research of the European wildcat (Felis silvestris) in Baden-Württemberg. Säugetierkundliche Informationen 8:411–416
Sunquist M, Sunquist F (2002) Wild cats of the world. University of Chicago Press, Chicago and London
Todesco M, Pascual MA, Owens GL, Ostevik KL, Moyers BT, Hubner S, Heredia SM, Hahn MA, Caseys C, Bock DG, Rieseberg LH (2016) Hybridization and extinction. Evol Appl 9:892–908. https://doi.org/10.1111/eva.12367
Valiere N (2002) GIMLET: a computer program for analysing genetic individual identification data. Mol Ecol Notes. https://doi.org/10.1046/j.1471-8286.2002.00228.x-i2
vonHoldt BM, Pollinger JP, Earl DA, Parker HG, Ostrander EA, Wayne RK (2013) Identification of recent hybridization between gray wolves and domesticated dogs by SNP genotyping. Mamm Genome 24:80–88. https://doi.org/10.1007/s00335-012-9432-0
von Thaden A, Cocchiararo B, Jarausch A, Jüngling H, Karamanlidis AA, Tiesmeyer A, Nowak C, Muñoz-Fuentes V (2017) Assessing SNP genotyping of noninvasively collected wildlife samples using microfluidic arrays. Sci Rep. https://doi.org/10.1038/s41598-017-10647-w
Yamaguchi N, Kitchener A, Driscoll C, Nussberger B (2015) Felis silvestris. In: The IUCN Red List of Threatened Species. https://dx.doi.org/10.2305/IUCN.UK.2015-2.RLTS.T60354712A50652361.en
Zwijacz-Kozica T, Wazna A, Muñoz-Fuentes V, Tiesmeyer A, Cichocki J, Nowak C (2017) Not European wildcats, but domestic cats inhabit Tatra National Park. Pol J Ecol 65:415–421
Acknowledgements
Open Access funding was provided by Projekt DEAL. Samples used in the frame of this study were collected over several years, and we warmly appreciate the help of numerous wildcat experts. In particular, we thank Thomas Mölich and Burkhard Vogel (BUND), Franz Müller, Manfred Trinzen, Annette Kohnen (FVA), Malte Götz, Gisbert Geisler, Ole Anders (Nationalpark Harz) and several federal conservation agencies (Thuringian State Department for Environment and Geology, Lower Saxon State Department for Waterway, Coastal and Nature Conservation, State Department for Environmental Protection Sachsen-Anhalt). We appreciate the technical support of several members of the Conservation Genetics group: Tobias Erik Reiners, Berardino Cocchiararo, Alina von Thaden, Hannah Jüngling, Yvonne Puder. AT received partial funding in the frame of the BUND-led project “Wildkatzensprung” funded by the German Federal Agency for Nature Conservation (BfN) with resources provided by the Federal Ministry for the Environment, Nature Conservation and Nuclear Safety (BMU). ACK thanks the Negaunee Foundation for its generous support of a curatorial preparator who prepared the samples from Scotland. MC thanks Luca Lapini (Friulian Museum of Natural History), Andrea Sforzi (Natural History Museum of Maremma) and Prof. Martin Fischer (Friedrich-Schiller University, Jena) for making the collection of the Italian samples possible. PM enjoyed a postdoctoral fellowship funded by FEDER funds through the Operational Programme for Competitiveness Factors—COMPETE, and by National Funds through FCT—Foundation for Science and Technology (UID/BIA/50027/2013 and POCI-01-0145-FEDER-006821).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tiesmeyer, A., Ramos, L., Manuel Lucas, J. et al. Range-wide patterns of human-mediated hybridisation in European wildcats. Conserv Genet 21, 247–260 (2020). https://doi.org/10.1007/s10592-019-01247-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-019-01247-4