Abstract
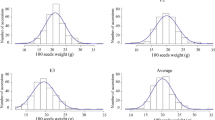
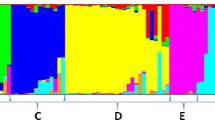
Seed-size traits, which are controlled by multiple genes in soybean, play an important role in determining seed yield, quality and appearance. However, the molecular mechanisms controlling the size of soybean seeds remain unclear, and little research has been done to investigate these mechanisms. In this study, we performed a genetic analysis to determine the genetic architecture of soybean seed size and shape via linkage and association analyses. We used 184 recombinant inbred lines (RILs) and 219 cultivated soybean accessions to evaluate seed length, seed width and seed height as seed-size traits, and their ratios of these values as seed-shape traits. Our results showed that all six traits had high heritability ranging from 92.46 to 98.47 %. Linkage analysis in the RILs identified 12 quantitative traits loci (QTLs), with five of these QTLs being associated with seed size, five with seed shape and two with the two first principal components of our principal component analysis (PCA). Association analysis in the 219 accessions detected 41 single nucleotide polymorphism (SNP)-trait associations, with 20 of these SNPs being associated with seed-size traits, seven with seed-shape traits and 14 with the two first principal components of our PCA. This analysis reveals that seed-size and seed-shape may be controlled by different genetic factors. Our results provide a greater understanding of phenotypic structure and genetic architecture of soybean seed, and the QTLs detected in this study form a basis for future fine mapping, quantitative trait gene cloning and molecular breeding in soybean.

Similar content being viewed by others
References
Ainsworth EA, Yendrek CR, Skoneczka JA, Long SP (2012) Accelerating yield potential in soybean: potential targets for biotechnological improvement. Plant Cell Environ 35(1):38–52
Atwell S, Huang YS, Vilhjálmsson BJ, Willems G, Horton M, Li Y, Meng D, Platt A, Tarone AM, Hu TT, Jiang R, Muliyati NW, Zhang X, Amer MA, Baxter I, Brachi B, Chory J, Dean C, Debieu M, Meaux JD, Ecker JR, Faure N, Kniskern JM, Jonathan DG, Jones G, Michael T, Nemri A, Roux F, Salt DE, Tang C, Todesco M, Traw MB, Weigel D, Marjoram P, Borevitz JO, Bergelson J, Nordborg M (2010) Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465:627–631
Bradbury P, Zhang Z, Kroon D, Casstevens T, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23(19):2633–2635
Cai H, Morishima H (2002) QTL clusters reflect character associations in wild and cultivated rice. Theor Appl Genet 104:1217–1228
Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138:963–971
Cober ER, Voldeng HD, Frégeau-Reid JA (1997) Heritability of seed shape and seed size in soybean. Crop Sci 37:1767–1769
Cockram J, White J, Zuluaga DL, Smith D, Comadran J, Macaulay M, Luo Z, Kearsey MJ, Werner P, Harrap D, Tapsell C, Liu H, Hedley PE, Stein N, Schulte D, Steuernagel B, Marshall DF, Thomas WTB, Ramsay L, Mackay I, Balding DJ, Consortium TG, Waugh R, O’Sullivan DM (2010) Genome-wide association mapping to candidate polymorphism resolution in the unsequenced barley genome. Proc Natl Acad Sci USA 107(50):21611–21616
Devlin B, Roeder K (1999) Genomic control for association studies. Biometrics 55(4):997–1004
Devlin B, Bacanu SA, Roeder K (2004) Genomic control to the extreme. Nat Genet 36:1129–1130
Famoso AN, Zhao K, Clark RT, Tung CW, Wright MH, Bustamante C, Kochian LV, McCouch SR (2011) Genetic architecture of aluminum tolerance in rice (Oryza sativa) determined through genome-wide association analysis and QTL mapping. PLoS Genet 7(8):e1002221. doi:10.1371/journal.pgen.1002221
Fan C, Xiong Y, Mao H, Lu T, Han B, Xu C, Li X, Zhang Q (2006) GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor Appl Genet 112:1164–1171
Feng X, Wilson Y, Bowers J, Kennaway R, Bangham A, Hannah A, Coen E, Hudson A (2009) Evolution of allometry in antirrhinum. Plant Cell 21:2999–3007
Flint-Garcia SA, Thornsberry JM, Bucker ES (2003) Structure of linkage disequilibrium in plants. Annu Rev Plant Biol 54:357–374
Frary A, Nesbitt TC, Grandillo S, Knaap E, Cong B, Liu J, Meller J, Elber R, Alpert KB, Tanksley SD (2000) fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science 289:85–88
Fu S, Zhan Y, Zhi H, Gai J, Yu D (2006) Mapping of SMV resistance gene Rsc-7 by SSR markers in soybean. Genetica 128:63–69
Gegas VC, Nazari A, Griffiths S, Simmonds J, Fish L, Orford S, Sayers L, Doonan LH, Snape JW (2010) A genetic framework for grain size and shape variation in wheat. Plant Cell 22:1046–1056
Hao D, Cheng H, Yin Z, Cui S, Zhang D, Wang H, Yu D (2012) Identification of single nucleotide polymorphisms and haplotypes associated with yield and yield components in soybean (Glycine max) landraces across multiple environments. Theor Appl Genet 124:447–458
Hayes AJ, Ma GR, Buss GR, Saghai Maroof MA (2000) Molecular marker mapping of Rsv4, a gene conferring resistance to all known strains of soybean mosaic virus. Crop Sci 40:1434–1437
Huang X, Wei X, Sang T, Zhao Q, Feng Q, Zhao Y, Li C, Zhu C, Lu T, Zhang Z, Li M, Fan D, Guo Y, Wang A, Wang L, Deng L, Li W, Lu Y, Weng QJ, Liu K, Huang T, Zhou T, Jing Y, Li W, Lin Z, Buckler ES, Qian Q, Zhang Q, Li J, Han B (2010) Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet 42:961–967
Hymowitz T (1970) On the domestication of soybean. Econ Bot 24:408–421
Jin J, Liu X, Wang G, Mi L, Shen Z, Chen X, Herbert SJ (2010) Agronomic and physiological contributions to the yield improvement of soybean cultivars released from 1950 to 2006 in Northeast China. Field Crops Res 115:116–123
Jofuku KD, Omidyar PK, Gee Z, Okamuro JK (2005) Control of seed mass and seed yield by the floral homeotic gene APETALA2. Proc Natl Acad Sci USA 102:3117–3122
Lam HM, Xu X, Liu X, Chen W, Yang G, Wong FL, Li MW, He W, Qin N, Wang B, Li J, Jian M, Wang J, Shao G, Wang J, Sun SM, Zhang G (2010) Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat Genet 42(12):1053–1059
Langlade NB, Feng X, Dransfield T, Copsey L, Hanna AI, Thebaud C, Bangham A, Hudson A, Coen E (2005) Evolution through genetically controlled allometry space. Proc Natl Acad Sci USA 102:10221–10226
Li Y, Huang Y, Bergelson J, Nordborg M, Borevitz JO (2010) Association mapping of local climate-sensitive quantitative trait loci in Arabidopsis thaliana. Proc Natl Acad Sci USA 107:21199–21204
Li X, Yan W, Agrama H, Jia L, Shen X, Jackson A, Moldenhauer K, Yeater K, McClung A, Wu D (2011a) Mapping QTLs for improving grain yield using the USDA rice mini-core collection. Planta 234:347–361
Li Y, Fan C, Xiong Y, Jiang Y, Luo L, Sun L, Shao D, Xu C, Li X, Xiao J, He Y, Zhang Q (2011b) Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat Genet 43:1266–1269
Liang HZ, Li WD, Wang H, Fang XJ (2005) Genetic effects on seed traits in soybean. Acta Genet Sin 32:1199–1204
Liang H, Wang S, Yu Y, Wang T, Gong P, Fang X, Liu X, Zhao S, Zhang M, Li W (2008) Mapping quantitative trait loci for six seed shape traits in soybean. Henan Agric Sci 45:54–60
Liu J, Van Eck J, Cong B, Tanksley SD (2002) A new class of regulatory genes underlying the cause of pear-shaped tomato fruit. Proc Natl Acad Sci USA 99:13302–13306
Lü HY, Liu XF, Wei SP, Zhang YM (2011) Epistatic association mapping in homozygous crop cultivars. PLoS One 6(3):e17773. doi:10.1371/journal.pone.0017773
Mao H, Sun S, Yao J, Wang C, Yu S, Xu C, Li X, Zhang Q (2010) Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc Natl Acad Sci USA 107:19579–19584
McCouch SR, Chen X, Panaud O, Temnykh S, Xu Y, Cho YG, Huang N, Ishii T, Blair M (1997) Microsatellite marker development, mapping and application in rice genetics and breeding. Plant Mol Biol 35:89–99
Morrison MJ, Voldeng HD, Cober ER (2000) Agronomic changes from 58 years of genetic improvement of short-season soybean cultivars in Canada. Agron J 92:780–784
Niu Y, Xu Y, Liu XF, Yang SX, Wei SP, Xie FT, Zhang YM (2013) Association mapping for seed size and shape traits in soybean cultivars. Mol Breed. doi:10.1007/s11032-012-9833-5
Ohto MA, Fischer RL, Goldberg RB, Nakamura K, Harada JJ (2005) Control of seed mass by APETALA2. Proc Natl Acad Sci USA 102(8):3123–3128
Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38:904–909
Qin H, Guo W, Zhang Y, Zhang T (2008) QTL mapping of yield and fiber traits based on a four-way cross population in Gossypium hirsutum L. Theor Appl Genet 117:883–894
Quarrie SA, Quarrie SP, Radosevic R, Rancic D, Kaminska A, Barnes JD, Leverington M, Ceoloni C, Dodig D (2006) Dissecting a wheat QTL for yield present in a range of environments: from the QTL to candidate genes. J Exp Bot 57:2627–2637
R Development Core Team (2010) R: a language and environment for statistical computing. R foundation for statistical Computing, Vienna
Raman H, Stodart B, Ryan PR, Delhaize E, Emebiri L, Raman R, Coombes N, Milgate A (2010) Genome-wide association analyses of common wheat (Triticum aestivum L.) germplasm identifies multiple loci for aluminium resistance. Genome 53:957–966
Rector BG, All JN, Parrott WA, Boerma HR (1999) Quantitative trait loci for antixenosis resistance to corn earworm in soybean.Crop Sci 39:531–538
Salas P, Oyarzo-Llaipen JC, Wang D, Chase K, Mansur L (2006) Genetic mapping of seed shape in three population of recombinant inbred lines of soybean (Glycine max L. Merr.). Theor Appl Genet 113:1459–1466
Schruff MC, Spielman M, Tiwari S, Adams S, Fenby N, Scott RJ (2006) The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 133:251–261
Shappley ZW, Jenkins JN, Zhu J, McCarty JC (1998) Quantitative trait loci associated with yield and fiber traits of upland cotton. J Cotton Sci 4:153–163
Singh AK, Fu D, El-Habbak M, Navarre D, Ghabrial S, Kachroo A (2011) Silencing genes encoding omega-3 fatty acid desaturase alters seed size and accumulation of Bean pod mottle virus in soybean. Mol Plant Microbe Interact 24(4):506–515
Song QJ, Mare LF, Shoemaker RC, Lark KG, Concibido VC, Delannay X, Specht JE, Cregan PB (2004) An new integrated genetic linkage map of the soybean. Theor Appl Genet 109:122–128
Song X, Huang W, Shi M, Zhu M, Lin H (2007) A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat Genet 39:623–630
Stoll M, Kwitek-Black AE, Cowley AW, Harris EL, Harrap SB, Krieger JE, Printz MP, Provoost AP, Sassard J, Jacob HJ (2000) New target regions for human hypertension via comparative genomics. Genome Res 10:473–482
Sun X, Shantharaj D, Kang X, Ni M (2010) Transcriptional and hormonal signaling control of Arabidopsis seed development. Curr Opin Plant Biol 13(5):611–620
van der Knaap E, Tanksley SD (2001) Identification and characterization of a novel locus controlling early fruit development in tomato. Theor Appl Genet 103:353–358
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93(1):77–78
Wang S, Basten C, Zeng Z (2005) Windows QTL cartographer version 2.5. Statistical genetics. North Carolina State University, Raleigh
Wang J, McClean PE, Lee R, Jay Goos R, Helms T (2008) Association mapping of iron deficiency chlorosis loci in soybean (Glycine max L. Merr.) advanced breeding lines. Theor Appl Genet 116:777–787
Wang A, Garcia D, Zhang H, Feng K, Chaud hury A, Berger F, Peacock WJ, Dennis ES, Luo M (2010) The VQ motif protein IKU1 regulates endosperm growth and seed size in Arabidopsis. Plant J 63:670–679
Wang S, Wu K, Yuan Q, Liu X, Liu Z, Lin X, Zeng R, Zhu H, Dong G, Qian Q, Zhang G, Fu X (2012) Control of grain size, shape and quality by OsSPL16 in rice. Nat Genet 44(8):950–954
Wen ZX, Zhao TJ, Zheng YZ, Liu SH, Wang CF, Wang F, Gai JY (2008) Association analysis of agronomic and quality traits with SSR markers in Glycine max and Glycin soja in China: 1. Population structure and associated markers. Acta Agron Sin 34(7):1169–1178
Weng J, Xie C, Hao Z, Wang J, Liu C, Li M, Zhang D, Bai L, Zhang S, Li X (2011) Genome-wide association study identifies candidate genes that affect plant height in Chinese elite maize (Zea mays L.) inbred lines. PLoS One 6(12):e29229. doi:10.1371/journal.pone.0029229
Wilson DO Jr (1994) Storage of orthodox seeds. In: Basra AS (ed) Seed quality: basic mechanisms and agricultural implications. Food Products Press, New York, pp 173–207
Xu Y, Li H, Li G, Wang X, Cheng L, Zhang Y (2011) Mapping quantitative trait loci for seed size traits in soybean (Glycine max L. Merr.). Theor Appl Genet 122:581–594
Yu J, Pressoir G, Briggs WH, Vroh BL, Doebley JF, McMullen MD, Gaut BS, Nielsen DM, Holland JB, Kresovich S, Buckler ES (2006) A unified mixed model for association mapping that accounts for multiple levels of relatedness. Nat Genet 38(2):203–208
Zhang J, Zhao TJ, Gai JY (2008) Association analysis of agronomic trait QTLs with SSR markers in released soybean cultivars. Acta Agron Sin 34(12):2059–2069
Zhou Y, Ni M (2010) SHORT HYPOCOTYL UNDER BLUE1 truncations and mutations alter its association with a signaling protein complex in Arabidopsis. Plant Cell 22:703–715
Zhou Y, Zhang X, Kang X, Zhao X, Zhang X, Ni M (2009) SHORT HYPOCOTYL UNDER BLUE1 associates with MINISEED3 and HAIKU2 promoters in vivo to regulate Arabidopsis seed development. Plant Cell 21:106–117
Acknowledgments
We are grateful to Dr. Hengyou Zhang for his assistance during the fieldwork and his advices in the manuscript. We thank the two anonymous reviewers for their valuable suggestions, which improved our manuscript. This work was supported by the National Basic Research Program of China (973 Program) (2010CB125906, 2009CB118400), the National Natural Science Foundation of China (31000718, 31171573, 31201230, 31271749), and the Jiangsu Provincial Programs (BE2012328, BK2012768, BE2012747).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hu, Z., Zhang, H., Kan, G. et al. Determination of the genetic architecture of seed size and shape via linkage and association analysis in soybean (Glycine max L. Merr.). Genetica 141, 247–254 (2013). https://doi.org/10.1007/s10709-013-9723-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-013-9723-8