Abstract
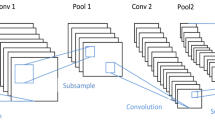
This paper addresses the demand for an intelligent and rapid classification system of skin cancer using contemporary highly-efficient deep convolutional neural network. In this paper, we mainly focus on the task of classifying the skin cancer using ECOC SVM, and deep convolutional neural network. RGB images of the skin cancers are collected from the Internet. Some collected images have noises such as other organs, and tools. These images are cropped to reduce the noise for better results. In this paper, an existing, and pre-trained AlexNet convolutional neural network model is used in extracting features. A ECOC SVM clasifier is utilized in classification the skin cancer. The results are obtained by executing a proposed algorithm with a total of 3753 images, which include four kinds of skin cancers images. The implementation result shows that maximum values of the average accuracy, sensitivity, and specificity are 95.1 (squamous cell carcinoma), 98.9 (actinic keratosis), 94.17 (squamous cell carcinoma), respectively. Minimum values of the average in these measures are 91.8 (basal cell carcinoma), 96.9 (Squamous cell carcinoma), and 90.74 (melanoma), respectively.








Similar content being viewed by others
References
Aldebaro K, Nikola J, Alon O (2003) On nearest-neighbor error-correcting output codes with application to all-pairs multiclass support vector machines. J Mach Learn Res 4:1–15
ali Bagheri M, Montazer GA, Escalera S (2012) Error Correcting Output Codes for multiclass classification: Application to two image vision problems. The 16th CSI International Symposium on Artificial Intelligence and Signal Processing (AISP 2012), p 508–513
Allwein EL, Schapire RE, Singer Y (2000) Reducing multiclass to binary: a unifying approach for margin classifiers. J Mach Learn Res 1:113–141
Andre E, Brett K, Novoa Roberto A, Justin K, Swetter Susan M, Blau Helen M, Sebastian T (2017) Dermatologist-level classification of skin cancer with deep neural networks. Nature 542:115–118
Blum A, Luedtke H, Ellwanger U, Schwabe R, Rassner G, Garbe C (2004) Digital image analysis for diagnosis of cutaneous melanoma. Development of a highly effective computer algorithm based on analysis of 837 melanocytic lesions. Br J Dermatol 151:1029–1038
Cireşan DC, Meier U, Masci J, Gambardella LM, Schmidhuber J (2011) Flexible, high performance convolutional neural networks for image classification. In: Proceedings of 22nd International Joint Conference on Artificial Intelligence (IJCAI ‘11) 22:1237–1242
Cireşan DC, Meier U, Schmidhuber J (2012) Multi-column deep neural networks for image classification. In: Proceedings of the CVPR, p 3642–3649
Daniel R, Vicente B, Antonio S, Belén S (2011) A decision support system for the diagnosis of melanoma: a comparative approach. Expert Syst Appl 38:15217–15223
Dietterich TG, Bakiri G (1995) Solving multiclass learning problems via error-correcting output codes. J Artif Intell Res 2:263–286
Ebtihal A, Arfan JM (2016) Classification of Dermoscopic skin cancer images using color and hybrid texture features. Int J Comput Sci Netw Secur 16(4):135–139
Germán C, Andrés C, Anabella B, Rodrigo A, Pablo M (2011) Toward a combined tool to assist dermatologists in melanoma detection from dermoscopic images of pigmented skin lesions. Pattern Recogn Lett 32:2187–2196
Hu W, Huang Y, Wei L, Zhang F, Li H (2015) Deep convolutional neural networks for Hyperspectral image classification. J Sensors 2015:258619
Ioannis G, Nynke M, Sander L, Michael B, Jonkman Marcel F, Nicolai P (2015) MED-NODE: a computer-assisted melanoma diagnosis system using non-dermoscopic images. Expert Syst Appl 42(19):6578–6585
Isasi AG, Zapirain GB, Zorrilla MA (2011) Melanomas non-invasive diagnosis application based on the ABCD rule and pattern recognition image processing algorithms. Comput Biol Med 41:742–755
Iyatomi H, Oka H, Celebi ME, Ogawa K, Argenziano G, Soyer HP, Koga H, Saida T, Ohara K, Tanaka M (2008) Computer-based classification of Dermoscopy images of melanocytic lesions on Acral volar skin. J Investig Dermatol 128:2049–2054
Krizhevsky A, Sutskever I, Hinton GE (2012) ImageNet Classification with Deep Convolutional Neural Networks. In: Proceedings of the Advances in Neural Information Processing Systems 25 (NIPS 2012) 1097–1105
Mohd A, Ram GK, Shafeeq A (2017) Skin cancer classification using K-means clustering. Int J Tech Res Appl 5(1):62–65
Na L, Xinbo Z, Yang Y, Xiaochun Z (2016) Objects classification by learning-based visual saliency model and convolutional neural network. Comput Intell Neurosci 2016:7942501
Qaisar A, Celebi ME, Carmen S, Fondón GI, Ma G (2013) Pattern classification of dermoscopy images: a perceptually uniform model. Pattern Recogn 46:86–97
Ramlakhan K, Shang Y (2011) A Mobile Automated Skin Lesion Classification System. In: Proceedings of 23rd IEEE International Conference on Tools with Artificial Intelligence, p 138–141
Sermanet P, Chintala S, LeCun Y (2012) Convolutional neural networks applied to house numbers digit classification. In: Proceedings of 21st International Conference on Pattern Recognition (ICPR ‘12), p 3288–3291
Sladojevic S, Arsenovic M, Anderla A, Culibrk D, Stefanovic D (2016) Deep neural networks based recognition of plant diseases by leaf image classification. Comput Intell Neurosci 2016:3289801
Somkid A, Jairaj P, Pawalai K (2011) Solving multiclass classification problems using combining complementary neural networks and error-correcting output codes. Int J Math Comput Simul 5(3):266–273
Vedaldi A, Lenc K (2016) MatConvNet: Convolutional Neural Networks for MATLAB. arXiv:1412.4564v3 [cs.CV], 5 May 2016 ml
Wiharto, Kusnanto H, Herianto (2015) Performance analysis of multiclass support vector machine classification for diagnosis of coronary heart diseases. Int J Comput Sci Appl 5(5):27–37
Xiao-Feng L, Xue-Ying Z, Ji-kang D (2010) Speech Recognition Based on Support Vector Machine and Error Correcting Output Codes. In: proceedings of the 1st International Conference on Pervasive Computing, Signal Processing and Applications, p 336–339
Yan Z, Yang Y (2014) Application of ECOC SVMs in Remote Sensing Image Classification. In: proceedings of the International Archives of the Photogrammetry, Remote Sensing and Spatial Information Sciences XL-2, ISPRS Technical Commission II Symposium, p 191–196
Yang Liu (2006) Using SVM and error-correcting codes for multiclass dialog act classification in meeting corpus. INTERSPEECH – ICSLP:1938–1941
Yushi C, Lin Z, Xing Z, Wang G, Yanfeng G (2014) Deep learning based classification of hyperspectral data. IEEE J Sel Top Appl Earth Observations Remote Sens 7(6):2094–2107
Zeiler MD, Fergus R (2014) Visualizing and understanding convolutional networks. ECCV 2014, Part I LNCS 8689:818–833
Zhigang Y, Yang Y (2014) Performance analysis and coding strategy of ECOC SVMs. Int J Grid Distrib Comput 7(1):67–76
Acknowledgements
In this research, Ulzii-Orshikh Dorj, Malrey Lee, and Keun-Kwang Lee conceived and designed the experiments; Ulzii-Orshikh Dorj, Keun-Kwang Lee performed the experiments; Ulzii-Orshikh Dorj, Keun-Kwang Lee, and Jae-Young Choi collected and analyzed the data; Ulzii-Orshikh Dorj, Keun-Kwang Lee contributed materials / analysis tools; Ulzii-Orshikh Dorj, Malrey Lee, Keun-Kwang Lee and Jae-Young Choi wrote the paper.
All named authors hereby declare that they have no conflicts of interest to disclose.
This work is supported by the National Research Foundation of Korea (NRF) granted by the Korea government (MSP) (No: 2017R1A2B4006667).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Dorj, UO., Lee, KK., Choi, JY. et al. The skin cancer classification using deep convolutional neural network. Multimed Tools Appl 77, 9909–9924 (2018). https://doi.org/10.1007/s11042-018-5714-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-018-5714-1