Abstract
The traction forces exerted by an adherent cell on a substrate have been studied only in the two-dimensions (2D) tangential to substrate surface (Txy). We developed a novel technique to measure the three-dimensional (3D) traction forces exerted by live bovine aortic endothelial cells (BAECs) on polyacrylamide deformable substrate. On 3D images acquired by confocal microscopy, displacements were determined with image-processing programs, and traction forces in tangential (XY) and normal (Z) directions were computed by finite element method (FEM). BAECs generated traction force in normal direction (Tz) with an order of magnitude comparable to Txy. Tz is upward at the cell edge and downward under the nucleus, changing continuously with a sign reversal between cell edge and nucleus edge. The method was evaluated regarding accuracy and precision of displacement measurements, effects of FE mesh size, displacement noises, and simple bootstrapping. These results provide new insights into cell-matrix interactions in terms of spatial and temporal variations in traction forces in 3D. This technique can be applied to study live cells to assess their biomechanical dynamics in conjunction with biochemical and functional activities, for investigating cellular functions in health and disease.
Similar content being viewed by others
Introduction
Cells communicate with the environment via mechanical forces as well as chemical signals. Cells are constantly subjected to mechanical forces generated internally or applied externally. These forces, which mediate the communication of cells with their neighboring cells and extracellular matrix (ECM), play an important role in the regulation of cell structure and function, including changes in adhesion sites and cytoskeletons, and alterations in cell motility, proliferation, differentiation, and survival.4 , 18 While mechanical forces can induce cellular responses,19 the cell can also modulate the mechanical forces it generates. Developments in traction force microscopy (TFM) have made it possible to determine the forces generated by cultured cells in tangential directions (X, Y) and to analyze the mechanical interplay between cells and their substrata.3 Harris et al. developed the first TFM by fabricating the deformable films made of silicone rubber.9 Several recent TFM techniques have been developed, including deformable substrate with embedded fluorescent beads,6 micro-patterned elastomer,2 micro-pillars,17 and micro-machined substrates.8
From the displacement field of the markers, the traction field can be computed from the known elastic material properties of the substratum containing the markers. Dembo and Wang formulated the traction force field from displacement field as a Boussinesq equation11 and then solved this inverse problem by regularization using Bayesian statistics.6 , 14 Butler et al. used Fourier transform to solve the Boussinesq equation.5 Schwarz et al. calculated the forces on each focal adhesion (FA) site with the a priori assumption that force is applied only on FAs.16 Regularization is applied to solve ill-conditioned Boussinesq equation as in Dembo and Wang. Yang et al. have applied Finite Element Analysis to remove the infinite half-plane assumption from the formulation of traction forces.20
Since cells are 3D in shape and can sense and respond to the 3D geometry in their environment,18 it is reasonable to assume that a cell can exert forces in all directions, but heretofore the plane stress assumption (i.e., with Tz = 0) was used in adherent cell studies to measure the cell traction force on the substratum. The current study was designed to assess this assumption by developing a novel method to determine the traction forces generated by a cell in the normal (Z) direction, in addition to those in the plane of the membrane (XY).
The present approach uses a combination of (a) a confocal microscope, (b) an image processing technique to determine 3D substrate deformation, and (c) a computational analysis to deduce 3D traction forces from substrate deformation. To calculate the displacements from the image, a particle-tracking algorithm based on the positions and volumes of the beads was used. We have used unconstrained approach in which all the displacements outside of the cell boundary are not removed in the calculation of traction forces. The advantages and disadvantages of unconstrained and constrained approaches have been discussed in a previous study.5 FEM was used to remove the substrate thickness effect from the traction force computation.
The properties of polyacrylamide (PAA) gel were evaluated in terms of elasticity and biocompatibility. We presented the evaluations of the methods through the displacement measurement, traction recovery simulation, effect of the noises on the traction, FE mesh size, and simple bootstrapping.
In conclusion, we have demonstrated that adherent BAECs exert 3D traction forces on the substrate. The application of this 3D TFM technique provides a new way to assess the full range of biomechanical dynamics of cells, which can be done in conjunction with their biochemical and functional activities, thus contributing to the understanding of cellular functions in health and disease.
Methods
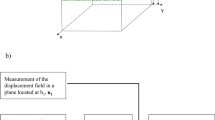
The present approach in determining cell traction forces is summarized in Fig. 1. It involves three main steps: (1) Experiments: Acquiring the 3D image stack, (2) Image Processing: Determining the displacement field from the image, and (3) Force Computation: Computing traction force field from the displacement field.
Conjugation of Fibronectin on Polyacrylamide Surface
PAA deformable substrate was prepared as described below, based on the method developed by Wang and colleagues.3 , 15 The 35 × 60 mm glass coverslip (Fisher Scientific International, Pittsburgh, PA) was activated with a Bunsen burner. Following the application of 0.1 M sodium hydroxide (Merck & Co., Inc., Whitehouse Station, NJ), the coverslip was siliconized with 3-aminopropyl triethoxy silane (Sigma–Aldrich Co., St. Louis, MO) followed by activation with 0.5% glutaraldehyde (Sigma).1 The acrylamide/bis-acrylamide solution was prepared with acrylamide (Bio-Rad Laboratories, Inc., Hercules, CA), N,N′-methylene bis-acrylamide (Bis, Bio-Rad), HEPES buffer, and distilled water (with final concentration of acrylamide/Bis = 5%/0.1%). 0.2-μm diameter red fluorescent (580/605) polystyrene beads (Invitrogen Corp., Carlsbad, CA) were added in the solution. After the addition of 0.06% ammonium persulfate (APS, Sigma) and 0.4% N,N,N′,N′-tetramethylene diamine (TEMED, Invitrogen), the solution was sandwiched and allowed to polymerize between the activated coverslip and unactivated coverslip of 22-mm diameter for 40 min at room temperature. After the polymerization, the unactivated coverslip was removed, and the bovine fibronectin (FN, Sigma) was cross-linked with PAA surface by activating N-sulfosuccinimidyl-6-[4′-azido-2′-nitrophenylamino] hexanoate (Sulfo-SANPAH, Pierce) with UV. UV with 252 nm wavelength was exposed twice for 7 min on the Sulfo-SANPAH solution on PAA. After the activation of Sulfo-SANPAH, 200 μL of 50 μg mL−1 FN solution in PBS was spread on the activated PAA substrate. The FN-conjugated PAA substrates were incubated overnight at 4 °C.
Determination of Mechanical Properties of the PAA Gel
To assess the elastic property of the PAA gel, the Young’s modulus (E) was determined with a compressive test device (V500cs, Biosyntech Inc., Laval, Québec, Canada). The 5% PAA specimens (diameter = 6 mm, thickness = 3 mm) with different concentrations of Bis were prepared using the same protocol as described above and kept in the cell culture medium. Stresses were measured by applying compressive strains from 0 to 25% to the PAA specimens, and E was calculated from the stress–strain relationship. The Young’s Modulus is shown in Supplementary Fig. 1 for an acrylamide concentration of 5%.
To assess the rheological properties of the gel, a compressive relaxation device was used to determine whether the PAA has a significant viscous component. For characterization of the PAA substrate, Student’s t-test (two-tailed) was used to analyze stress relaxation by comparing the stresses before and after 1-h relaxation. The null hypothesis of no significant time-dependence was accepted with a significance level of 0.05. The PAA gel (within the range of Bis concentrations used) is assumed to be an isotropic elastic material based on the results in stress relaxation tests. Thus, Young’s modulus (E) and Poisson’s ratio (ν) are the only two material variables needed for the formulation of the governing equations. Our results on the measurement of E are similar to those previously reported by others using atomic force microscopy (AFM) and tensile test,7 despite the differences in geometry and the surface of the substrate, as well as the methods used (see Supplementary Fig. 1). The value of 0.3 was used as the Poisson’s ratio, as measured in a previous study.12
Cell Culture
Cell culture reagents were purchased from Invitrogen unless otherwise mentioned. BAECs were isolated from the bovine aorta obtained from Cal Poly University, Pomona (Pomona, CA, http://www.csupomona.edu/~meatlab/) with the use of collagenase.13 The cells were cultured in a 10-cm Petri dish containing Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% sodium pyruvate, 1% l-glutamine, and 1% penicillin–streptomycin. Culture was maintained in a humidified 5% CO2/95% air incubator at 37 °C. Experiments were conducted with cells before passage 15.
Verification of Biocompatibility of Substrate
Although PAA is generally considered to be inert to cells, any remaining unpolymerized monomer, initiator or catalyst molecules might be toxic.
In order to assess the biocompatibility of the PAA substrate, 0.2% trypan blue solution (Invitrogen) in PBS was added to the BAECs that had been cultured overnight on the PAA substrate and allowed to incubate for 10 min. After washing once with PBS, the presence of BAECs stained with trypan blue was determined under the microscope. Trypan blue test showed no dye uptake by BAECs after the overnight incubation. Thus, the cells maintained their viability on the substrate, indicating that the PAA gels used in the experiments are biocompatible.
Microscopy and Image Enhancement
FN-conjugated PAA substrates were exposed to UV light for 10 min to minimize microbial contamination. BAECs were allowed to attach onto the FN-conjugated substrate overnight in the cell culture media. During the experiment, live BAECs were kept in a rectangular flow channel (0.1 cm in height, 2.3 cm in width, and 5 cm in length) formed by sandwiching a silicone gasket between the 35 × 60 mm glass slide with the PAA substrate and a glass plate attached onto an aluminum housing. The chamber was kept at 37 °C with a temperature control system.
We used spinning disk confocal microscopy and image enhancement setup (3D deconvolution) to guarantee the high resolutions for determining displacement and traction. The Z-stack was obtained by imaging a XY plane and adjusting the stage height by 0.2 microns between each plane. It took less than 3 min to acquire the Z-stack, including the saving time onto the hard disk. The spinning disk confocal microscope (IX-81, Olympus America Inc., Center Valley, PA) with a 60× objective lens (UIS Plan-Apo, N.A. 1.40, Olympus) was used to track the motions of the embedded red fluorescent (580/605) beads (fluorescence mode) and BAECs (DIC mode) The magnification scale was 0.1075 μm/pixel in X, Y directions, and the optical section step was 0.2 μm. “Force-loaded” images were acquired from the bead-embedded gel on which BAECs were attached and exerted their traction force. The “Null-force” images were taken from the gel without the BAECs following their removal with trypsin to eliminate the cell-induced gel deformation. After the image acquisition, Meta-Morph 6.3 (Molecular Devices Corp., Downingtown, PA) was used for the image-enhancement post-processing step, including background subtraction, 2D-deconvolution, and median filtering. AutoQuant 9 (Media Cybernetics Inc., Bethesda, MD) was used for the 3D-deconvolution.
Determination of the Threshold Value and Bead Coordinates
A MATLAB-coded program was used to obtain the 3D coordinates of fluorescent beads. In order to separate the fluorescence intensities between the object (bead) and the background during the image segmentation procedure, a threshold value needs be established to cutoff the background pixel values. We determined this threshold value on a physical basis by using the known marker bead concentration measured with spectrophotometry.
A range of cutoff values (candidates of the threshold value) were applied to the program coded in MATLAB, and the image-processed apparent bead concentrations were obtained accordingly. From the measured known concentration of 5.36 × 1010 beads mL−1, the threshold value was determined to be 0.023 (Fig. 2a). The plot of image-processed apparent bead concentrations against cutoff values exhibits a plateau region in which the apparent bead concentration does not vary significantly. This is because of the marked differences between the signals from the beads and those due to background noises (Fig. 2b). The physically determined threshold value lies in this plateau region. After binarization, the 3D coordinates of the center of mass (centroid) and the volumes of the beads can be calculated.
Determination of threshold value. (a) Determination of threshold value from the apparent bead concentrations measured at different cutoff values, Dots: Image-processed apparent bead concentrations obtained from different cutoff values as candidates for determining the threshold value, Square: Bead concentration measured experimentally by spectrometry (5.36 × 1010 mL−1), with a corresponding cutoff value of 0.023 on the abscissa. The intensity values of the bead-containing gels were plotted on a relative scale between 0 and 1. (b) Schematic diagram showing the principle of binarization (thresholding), Solid curve: 16-bit grayscale image (normalized from 0 to 1), Dashed line: 1-bit binary image, Dotted horizontal line: threshold value
Determination of 3D Displacements Using Volume Comparison
A program of particle tracking algorithm (PTA) was coded in MATLAB to track the displacements of given beads from each null-force bead set {B 0} to the force-loaded bead set {B 1}. The displacement was determined from the measured coordinate changes, taking advantage of the characteristic volume of each bead.
The program involves two steps: First, we found the five close-neighbor beads (bead1’s) in set {B 1} for one given bead (bead0) in set {B 0}, i.e., the five bead1’s that had the closest distances from bead0. We set the maximum limit of the distance (d max) between the bead0 and the bead1’s as 1.5 μm because the measured maximum displacement of the bead was about 1.2 μm. Then, the volume of each of these five bead1’s (V 1’s) was compared with that of bead0 (V 0). The bead1 with the smallest volume difference from bead0 was selected as the same bead as bead0, and the distance between the two was taken as the displacement distance. This five-neighbors and nearest-volume (5NV) algorithm was used to determine the displacement of each bead from the coordinates at each time point.
In order to correct for any possible displacement field due to microscope stage drift, we determined the displacements of beads >15 μm away from the cell, where the influence of the cell traction is negligible, and subtracted these from the displacement field at each node before the traction force computation.
Because the beads (or data points) are randomly distributed in the PAA gel, a three-dimensional linear interpolation is applied to obtain the displacements at desired nodes as inputs for FEM computation.
Determination of the Substrate Thickness
We tracked the Z-directional brightness of the top-most beads in the gel to determine the maxima, which correspond to the bead centers. The Z-coordinate of the top surface was determined as one radius (0.1 μm) above the Z-coordinate of these maxima. The Z-coordinate of the bottom surface was similarly determined. The substrate thickness was calculated from the Z-coordinates of the top and bottom surfaces. Gels of 18-27 μm thickness were casted by using 6–9 μL of pre-polymer solution. The measured thickness value was used in the traction force computation.
The substrate thickness is a function of the temperature and osmolality of its aqueous environment in the chamber. The temperature was maintained at 37 °C with a control system. The osmolality at 37 °C was 299 mOsm kg−1 for both the cell culture media (used before cell removal) and the PBS (used after cell removal). The substrate thickness before and after the cell removal was measured to be the same.
Computation of Traction Forces Based on FEM
In our computation, the PAA substrate on glass is assumed to be a plate with a finite thickness h. The schematic diagram in which a migrating cell exerts traction forces on the underlying deformable substrate is shown in Fig. 3. The PAA block (gray block under the cell) is the domain for the traction force analysis.
Schematic diagram showing a cell applying forces on the deformable substrate. (a) A cell exerts traction forces onto underlying PAA substrate. (b) Deformable substrate with cell traction forces. The PAA substrate is attached on the glass surface. W, H, and h: dimensions of PAA domain for the analyses in X-, Y-, and Z-directions, respectively
Isotropic and elastic material properties are assumed based on the characterization of mechanical properties mentioned above. The governing equations for the 3D boundary value problem (BVP) and boundary conditions (BCs) are as follows.
The strain–displacement relationship is
where ε ij = Strain tensor (i, j = 1, 2, 3), u i = Displacement vector, and x i = Right-handed Cartesian coordinates (i = 1, 2, 3).
The elastic stress–strain law is
where σ ij = Stress tensor, E = Young’s modulus, ν = Poisson’s ratio, δ ij = Kroneker delta.
The equation of static equilibrium for stresses inside of the domain is
The boundary conditions on displacement and stress are as follows:
-
u i = u * i at top plane (interface plane between cell and gel)
-
u i = 0 at bottom plane (interface plane between gel and glass)
-
σ ij n i = 0 otherwise (at the side planes)
where u * i = Measured displacement at the top surface between the cell and PAA, n j = Surface normal vector at the side planes (planes except top and bottom).
Equations (1), (2), and (3) form a static differential equation of 3D Hookean solid between stress and displacement, together with boundary conditions. We applied the FEM to solve the stress field (σ ij ) from the displacement field (u i ). Generally, finite element (FE) formulation turns the differential equation into an integration form and eventually into a matrix form Ku = f, where u is a nodal displacement vector, f a nodal force vector, and K a global stiffness matrix. With the boundary nodal force acquired from u * i , nodal displacement can be solved from the linear algebraic equations; thus the displacement field and stress field can be determined in the whole domain. Cell traction force can be calculated from the stress field of the top plane.
For FE analysis, the hexahedron (brick) type element, which has 8 nodes at the corner and eight Gauss integration points inside, was used. Two-micrometers fixed mesh grids were used as ΔX and ΔY. In the Z direction, ΔZ varies from 0.5 μm near the surface to 8 μm near the bottom of the gel. Thus, finer ΔZ was used as Z got close to cell-attached plane.
ABAQUS 6.5 (SIMULIA, Providence, RI), a commercial software package for FE analysis, was used to solve BVP.
Evaluation of the Force Computation Method
The inversion step, in which the stresses are calculated from the displacements, could adversely affect the accuracy, stability, and convergence of the solution. Hence, we evaluated the traction force computation method though traction force recovery, the effects of displacement noises and FE size, and simple bootstrapping. The Young’s Modulus of the material was 3.78 kPa, which is the same as in cell experiments.
Simulation: Traction Force Recovery
To assess how accurately the traction field can be recovered in our FEM method, we have simulated the traction force recovery using the following steps. First, we computed a displacement field from the known traction field prepared from four balanced distributed loads of 2.0 nN on the area of 4 μm2 each (Fig. 4a). Second, this displacement field was used as an input to calculate the traction field (Fig. 4b). Then, the traction fields at step one and step two were compared (Fig. 4c). U is the magnitude of displacement, and T is the magnitude of traction.
Simulation of traction force recovery and introduction of displacement noises. (a) Four balanced forces for the generation of displacement fields. (b) Calculated displacement field from the traction fields. (c) Contour plot of traction difference (||T(recovered) − T(input)||). (d) Effects of the introduced Gaussian noises of 0, 0.01, 0.05, 0.1, and 0.2 μm on the RMS of traction force error (ΔT = ||T(with noise) − T(without noise)||). The R 2 value was 0.999. U = magnitude of displacement, and T = magnitude of traction
Simulation: Effect of Displacement Noises
To evaluate the stability of our computational system, a simulation was performed to show the effect of displacement noises on the traction solution. If the system is stable (or K is well-conditioned), traction errors are small, but if it is unstable (or K is ill-conditioned), traction errors are amplified for the same input (displacement) noises. We introduced random Gaussian noises in 3D with standard deviations (SDs) of 0 (control), 0.01, 0.05, 0.1, and 0.2 μm in each nodal boundary displacement field. Traction fields were calculated from these displacements fields, and the root mean square (RMS) differences of traction forces obtained with (0.01, 0.05, 0.1, and 0.2 μm) and without (0 μm) displacement noises. We calculated the nodal displacements by interpolation from randomly distributed bead measurements. Strictly speaking, the simulated noise should be applied to these simulated bead measurements before interpolation onto a grid. Since our main focus here was to demonstrate the validity of the FEM method by the ABAQUS solver, we have used a simpler approach for the simulation.
Simulation: Effect of FE Mesh Size
If FE mesh size is not small enough, the result is not accurate, although converged solution can be achieved. Thus, mesh sets with finer ΔX and ΔY (ΔX = ΔY, 0.5, 1.0, and 2.0 μm) were prepared, and the traction forces computed with the simulated displacement input were compared.
Simple Bootstrapping of Cell Traction Force
To assess the level of the tractions with significance, simple bootstrapping16 was applied to the cell traction data. The measured SD of displacement (0.035 μm) was used as noise input (SD of Gaussian noises at each node). After the iteration (n = 10) of traction calculations, the SD of the traction calculated at each node was used as filtering criteria, i.e., the traction at a node was considered to be significant if it was greater than the SD at the node.
Results
Evaluation of the Method
Accuracy and Precision of Displacement Measurements
The displacements of beads located between the bottom and 1 μm above the bottom of the substrate should be essentially zero because the glass coverslip underneath prohibits the deformation of substrate. Displacements are obtained after the rigid body motion correction (or the registration correction). The closeness of the mean value (1.20 × 10−16 μm) of the measured displacement histogram of beads in this layer to the reference value 0 shows the high accuracy, while the small SD (0.035 μm) shows a high precision. This small dispersion (SD) around 0 displacement can be attributed to the small errors originated from the experiment (image acquisition, CCD digitization) and image processing (image enhancement, segmentation).
Traction Force Recovery
Traction field was reconstructed from the displacement field calculated from the balanced force setting as in Fig. 4a. The displacement calculated from the input traction field is shown in Fig. 4b. The traction difference (ΔT = ||T(recovered) − T(input)||) are shown in Fig. 4c. The RMS of the difference was found to be 9.6 × 10−6 kPa, which is less than 0.01% of the maximum traction. This traction recovery simulation verifies the accuracy of our traction force calculation. Detailed results in tangential and normal directions are shown in Supplementary Fig. 2.
Effect of Displacement Noises
Although we have shown the accuracy of our traction force calculation, it is worthwhile to evaluate the stability of our solution (traction) as mentioned above. In the simulation of traction forces with artificial Gaussian noises, the effect of noises introduced in the displacement to the RMS differences of traction is shown in Fig. 4d. The signal/noise ratios (maximum displacement/imposed noise) were 18.1, 3.6, 1.8, and 0.9 for noises of 0.01, 0.05, 0.10, and 0.2 μm respectively.
As we are dealing with a linear system, the errors and RMS of traction force error (ΔT = ||T(with noise) − T(without noise)||) show linear behavior with R 2 value of 0.9999.
Mesh Size Effect, Simple Bootstrapping, and Global Force Summation
The RMS difference of traction between the 0.5 and 2.0 μm meshes was 0.003 kPa and that between 0.5 and 1.0 μm was 0.0023 kPa. The maximum difference of traction between the 0.5 and 2.0 μm mesh was about 0.005 kPa and that between 0.5 and 1.0 μm was about 0.009 kPa. These results support the validity of the mesh size used (2.0 μm). The pattern of overall traction force differences is shown in Supplementary Fig. 3.
Simple bootstrapping was applied to the cell traction data, and the RMS of traction SDs was found to be about 0.043 kPa. The original traction force and the traction force after bootstrapping did not show significant differences.
The equation of global force summation at the top plane is as follows.
The measured global force sum was F = [Fx Fy Fz] = [0.56, 0.56, −5.8] nN, while the average traction forces in each direction was 〈T〉 = [〈Tx〉 〈Ty〉 〈Tz〉] = [0.00034, 0.00024, −0.0035] kPa, respectively.
Traction Force Field by BAECs
Marker Movements as Evidence of 3D Displacement Field
The 3D displacements of the substrate are presented in the top and side-cut views of the reconstructed fluorescence images (Fig. 5). Pseudocolors were applied to the TRITC-labeled beads when BAECs were present (green: force-loaded) and absent (red: null-force), and the images were merged. Thus, the direction of the bead movement (or force) was from red to green, and the yellow-colored beads indicate that they were not moved by the BAEC.
3D movements of the marker beads. (a) Top view of fluorescent beads at the top surface of the substrate, White contour lines: cell boundary and nucleus outline, (b) top view of fluorescent beads at the bottom surface of the substrate, (c) top view of BAEC with DIC at the top surface, with circles showing different regions corresponding to those shown in (d), (e), and (f), respectively. A: under the nucleus, B: under the cell edge, C: between nucleus edge and cell edge. There is no movement in XY plane for A. (d), (e), and (f): Top views showing the XY movements for A (no movement), B and C, respectively, as well as the section lines that generated the side-cut views to display the 3D movements of the beads at positions A, B, and C in (g), (h), and (i), respectively. Arrows adjacent to A, B and C show the direction of displacements of the marker beads. Bars = 10 μm in a, b, c, d, e, and f, and 5 μm in g, h and i, red: beads without BAEC, green: beads with BAECs, yellow: superimposition of red and green beads (no bead movement), Uxy = Tangential displacement. Uz = Normal displacement in Z-direction. Uz < 0 means downward, and Uz > 0 upward
Tangential movements of the beads at the top and bottom of the substrate are shown in Figs. 5a and 5b, respectively. The direction of tangential displacements (Uxy, from red to green) of beads near the substrate top is generally toward the cell center. Beads far away from the cell show no movement (yellow). Large Uxy values are concentrated at the cell edge, whereas small Uxy values are observed under the nucleus (Fig. 5a). No movement of the marker beads is observed at the substrate bottom (Fig. 5b).
Figure 5c shows the DIC image of the BAEC with three regions of interest at the top surface of the substrate: (A) under the nucleus, (B) under the cell edge, and (C) in between, depicting movements in the Z- as well as X- and Y-directions. Bead movements are shown in top view (XY-view in Figs. 5d–5f), and side-cut view (Z-view in Figs. 5g–5i). Under the nucleus, bead movements are purely downward with Uz < 0 (Fig. 5g) and Uxy ≅ 0 (Fig. 5d). At the cell edge, Z-direction displacements are upward with Uz > 0 (Fig. 5h), and Uxy is large (Fig. 5e). Between the nucleus and cell edge, displacement undergoes transition from being downward (Uz < 0) to upward (Uz > 0) (Fig. 2i), and Uxy is significant but smaller than that at the cell edge (Fig. 5f).
Figures 5h and 5i show both tangential and normal movements of the marker beads in the same image.
3D Displacement Field
The deformation of the PAA substrate by a BAEC was reconstructed from the displacements of marker beads, and is shown in Fig. 6 as a whole view (a) and a side-cut view (b). The contours of Uxy (c) and Uz (d) are shown at the top surface. The substrate is pulled up at the cell edge (red) and pushed down under the cell nucleus (blue). The bottom of the substrate, as expected, does not undergo any displacement (Figs. 6a and 6b).
3D displacement field and traction force field induced by BAEC. (a) Whole view (mainly top view) of the substrate. (b) Side-cut view of the substrate at Y = 32 μm. Mesh size in X, Y directions = 2 μm, Thickness of the deformable substrate = 17 μm. Young’s modulus E = 3.78 kPa. (c) Contour of tangential displacement (Uxy). Arrows indicate the directions of displacements. (d) Contour of normal displacement (Uz). Uz > 0 is upward and Uz < 0 downward. (e) Contour of Txy. Arrows indicate the direction of Txy. (f) Contour of Tz. Tz > 0 is upward and Tz < 0 downward. Dark lines in (a), (c), (d), (e) and (f): cell boundary and nucleus outline of BAEC
3D Traction Force Field
The 3D traction forces exerted by the BAEC were computed from the displacements. Figures 6e and 6f show the contours of Txy and Tz, respectively, at the top surface. The traction force patterns are similar to those of displacement. Txy is large at the cell edge and small under the nucleus (Fig. 6e). Tz is upward at the cell edge and downward under the nucleus (Figs. 6f and 7b), as in the case of displacements (Fig. 6d). The values of Tz change continuously and undergo a reversal of sign between the cell edge and the nucleus edge. The order of magnitude of the largest ||Tz|| value (Tz max) was comparable to those of ||Tx|| and ||Ty|| (Tx max and Ty max), while Tx max, Ty max, and Tz max were 0.74, 0.75, and 0.43 kPa, respectively.
Statistics of normal traction force. (a) Schematic diagram of subcellular categorization for the Tz analysis in (b). The results in the nucleus (inside the nucleus edge) are averaged and designated as “Nucleus”. To represent the data in region from the nucleus edge to the cell edge in annular rings, radial lines are drawn from the center of the nucleus at angles of 0°, 45°, 90°, 135°, 180°, 225°, 270°, and 315°, and each line is divided into three equal segments with four points (cell edge, two intermediate points, and nuclear edge). Connection of the corresponding points in circumferential direction results in the formation of four annular subregions from the cell edge to the nuclear edge. Data obtained from these annular subregions are referred to as Cell_edge, Cyto1, Cyto2, and Nuc_Edge. (b) Change of Tz across the cell from the cell edge to the nucleus. p-Value is from one-way analysis of variance (ANOVA). # p < 1.0 × 10−5, *p < 0.01 compared with Nucleus. Bar: standard deviation, N = 5 cells
Statistics
One-way analysis of variance (ANOVA) was used to test the significance of difference of the Tz values across the cell (N = 5 cells). The variation of Tz as a function of distance from the cell edge to the nucleus has a p-value = 3.46 × 10−6. Protected multi-comparison tests with Tukey’s honestly significant difference criterion10 after the ANOVA show a significance level of 1.0 × 10−5 for the difference of Tz between cell edge (Cell_Edge) and nucleus (Nucleus) (# in Fig. 7b) and 0.01 for the difference of Tz between Nucleus and Cyto1 (* in Fig. 7b).
Mesh Size Effect
The RMS difference of traction between the 0.5 and 1.0 μm meshes was 0.03 kPa and that between 0.5 and 2.0 μm was 0.04 kPa. Mesh size effect was larger with the cell traction data compared to that of simulation mentioned above. The maximum difference of traction between the 0.5 and 2.0 μm mesh was about 0.4 kPa and that between 0.5 and 1.0 μm was about 0.3 kPa. The pattern of overall traction force differences is shown in Supplementary Fig. 4.
Discussion
In the research presented in this article, a novel technique has been developed to determine the traction force exerted by a cell in the Z direction perpendicular to the substrate surface, as well as in the X, Y tangential directions. With this new technique, we have demonstrated that live cells on a deformable substrate exert forces in 3D rather than only 2D. Since the shape of the cell is nearly flat, previous researchers have neglected Tz of the traction forces. Our study shows that when the cell is situated on a flat surface, which is usually the condition of adherent cells, e.g., in culture, the cell edge experiences an upward or pulling up force, whereas the cell nucleus experiences a downward or pushing down force. One of the advantages of our method based on FEM and not on the Boussinesq equation is the removal of the infinite half-plane assumption. Cells on a substrate with finite thickness experience smaller displacement than those on infinite thickness. It has been demonstrated that the finite thickness effect cannot be neglected for the range of thickness generally used in traction experiments.20 Since our BVP formulation utilizes the information at the top region only, 3D scan of the whole gel is not necessary, thus resulting in an enhancement of time resolution. The periodic boundary condition of the FFT-based method is also unnecessary because our calculation is in the spatial domain.
The converging speed of our solution is very fast without the additional regularization step, so that force computation with 15,129 nodes could be completed within 5 min with a general purpose PC (Intel Core™2, 2.33 GHz CPU, 2.0 GB of RAM) and implicit solver.
The validity of mesh size (2.0 μm, ΔX = ΔY) for the FEM computation was demonstrated by comparing the results and those computed with finer meshes (0.5 and 1.0 μm, ΔX = ΔY), as little differences were shown among three meshes.
We have used an unconstrained method because the cell boundary information needs not be included, and hence the computation does not require investigator’s judgments. We recognize, however, that both constrained and unconstrained methods have advantages and disadvantages.5
The accuracy of the calculation was tested by simulation of traction force recovery, which demonstrates the validity of our traction force computation method. Traction errors show a linear behavior with artificially introduced displacement errors (Fig. 4d), with R 2 value close to 1. This plot can be used to estimate traction resolution from measuring the displacement resolution. If the slope of the plot is large, traction resolution becomes higher with the same displacement resolution. The main factor affecting the slope is the Young’s modulus E of the substrate: an increase in E would increase the slope. Hence, measuring the traction force on a rigid substrate may lead to an decrease of resolving power in traction. This may not present a problem if the traction force of interest is larger than this traction resolution, because there would be a sufficient signal (traction) to noise (traction resolution) ratio. But if the traction force of interest is small, the E effect would become important.
The voxel dimension of the 3D image is 0.1075 × 0.1075 × 0.2 μm, i.e., image pixel size of X and Y was 0.1075 μm, while the scan size of Z was 0.2 μm. Z step size could be reduced to enhance the displacement resolution and traction resolution in Z, but then we will have to sacrifice the time resolution because the Z-scan step is the time-limiting step of the experiment. It is also to be noted that long-term exposure to fluorescent light during the Z scan could induce the problem of photo-bleaching.
As Tx max, Ty max, and Tz max were 0.75, 0.76, and 0.43 kPa, the ratio Tz max/Tx max or Tz max/Ty max were about 0.6. Thus, Tz cannot be neglected.
Conclusions
We have developed a novel technique to measure the 3D traction forces (including Tz) exerted by live cells by using the PAA deformable substrate method. The substrate was characterized, including its elasticity, biocompatibility, and marker bead concentration. We acquired 3D images by using a confocal microscopy, and developed image-processing programs coded in MATLAB to obtain the displacement fields from the 3D image stack. We have also developed an improved 3D approach to compute the traction forces based on FEM. This method is effective in that it gives fast converging solutions with less demand on computational power. We have evaluated the method in terms of accuracy and precision of displacement measurements, simulation of traction force recovery, simulation of displacement noises effect, assessment of effects of FE mesh size, and simple bootstrapping. This is the first study that allows the measurement of traction force in the normal direction by using the traction force microscopy technique.
We have applied this new method to determine 3D traction forces exerted by live BAECs on the substrate, including the normal component, as a function of time. Thus, this method allows the study of cell dynamics in 4D (3D in space plus time). This traction force microscopy technique provides a new way of elucidating the full range of biomechanical dynamics of cells in conjunction with their biochemical and functional activities and can contribute to the understanding of cellular functions in health and disease.
References
Aplin, J. D., and R. C. Hughes. Protein-derivatised glass coverslips for the study of cell-to substratum adhesion. Anal. Biochem. 113(1):144–148, 1981.
Balaban, N. Q., U. S. Schwarz, D. Riveline, P. Goichberg, G. Tzur, I. Sabanay, D. Mahalu, S. Safran, A. Bershadsky, L. Addadi, et al. Force and focal adhesion assembly: a close relationship studied using elastic micropatterned substrates. Nat. Cell Biol. 3(5):466–472, 2001.
Beningo, K. A., C. M. Lo, and Y. L. Wang. Flexible polyacrylamide substrata for the analysis of mechanical interactions at cell-substratum adhesions. Methods Cell Biol. 69:325–339, 2002.
Bershadsky, A. D., N. Q. Balaban, and B. Geiger. Adhesion-dependent cell mechanosensitivity. Annu. Rev. Cell Dev. Biol. 19:677–695, 2003.
Butler, J. P., I. M. Tolic-Norrelykke, B. Fabry, and J. J. Fredberg. Traction fields, moments, and strain energy that cells exert on their surroundings. Am. J. Physiol. Cell Physiol. 282(3):C595–C605, 2002.
Dembo, M., and Y. L. Wang. Stresses at the cell-to-substrate interface during locomotion of fibroblasts. Biophys. J. 76(4):2307–2316, 1999.
Engler, A., L. Bacakova, C. Newman, A. Hategan, M. Griffin, and D. Discher. Substrate compliance versus ligand density in cell on gel responses. Biophys. J. 86(1 Pt 1):617–628, 2004.
Galbraith, C. G., and M. P. Sheetz. A micromachined device provides a new bend on fibroblast traction forces. Proc. Natl Acad. Sci. USA 94(17):9114–9118, 1997.
Harris, A. K., P. Wild, and D. Stopak. Silicone rubber substrata: a new wrinkle in the study of cell locomotion. Science 208(4440):177–179, 1980.
Hochberg, Y., and A. C. Tamhane. Multiple Comparison Procedures, Vol. xxii. New York: Wiley, p. 450, 1987.
Landau, L. D., E. M. Lifshitz, A. M. Kosevich, and L. P. Pitaevskiæi. Theory of Elasticity, Vol. viii. Oxford, Boston: Butterworth-Heinemann, p. 187, 1995.
Li, C., Z. Hu, and Y. Li. Poisson’s ratio in polymer gels near the phase-transition point. Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Topics 48(1):603–606, 1993.
Li, S., B. P. Chen, N. Azuma, Y. L. Hu, S. Z. Wu, B. E. Sumpio, J. Y. Shyy, and S. Chien. Distinct roles for the small GTPases Cdc42 and Rho in endothelial responses to shear stress. J. Clin. Invest. 103(8):1141–1150, 1999.
Marganski, W. A., M. Dembo, and Y. L. Wang. Measurements of cell-generated deformations on flexible substrata using correlation-based optical flow. Methods Enzymol. 361:197–211, 2003.
Pelham, Jr., R. J., and Y. Wang. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc. Natl Acad. Sci. USA 94(25):13661–13665, 1997.
Schwarz, U. S., N. Q. Balaban, D. Riveline, A. Bershadsky, B. Geiger, and S. A. Safran. Calculation of forces at focal adhesions from elastic substrate data: the effect of localized force and the need for regularization. Biophys. J. 83(3):1380–1394, 2002.
Tan, J. L., J. Tien, D. M. Pirone, D. S. Gray, K. Bhadriraju, and C. S. Chen. Cells lying on a bed of microneedles: an approach to isolate mechanical force. Proc. Natl Acad. Sci. USA 100(4):1484–1489, 2003.
Vogel, V., and M. Sheetz. Local force and geometry sensing regulate cell functions. Nat. Rev. Mol. Cell Biol. 7(4):265–275, 2006.
Wang, Y., E. L. Botvinick, Y. Zhao, M. W. Berns, S. Usami, R. Y. Tsien, and S. Chien. Visualizing the mechanical activation of Src. Nature 434(7036):1040–1045, 2005.
Yang, Z., J. S. Lin, J. Chen, and J. H. Wang. Determining substrate displacement and cell traction fields—a new approach. J. Theor. Biol. 242(3):607–616, 2006.
Acknowledgments
We wish to thank Dr. Shunichi Usami for advice and help on live cell chamber design, Phu Nyugen for assisting in the cell culture work, Gerard Norwich for help on microscopy, and Dr. Juan Carlos Del Alamo for advice on the simulations. This work was supported in part by National Institutes of Health research grants HL080518 and HL085159.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1
Elastic property of polyacrylamide gel. Young’s modulus of polyacrylamide gel determined by compressive test vs. bis-acrylamide (cross-linker) concentration (Bis = 0.05–0.6 w/v%), Bars: standard deviation. Inset: Data for Bis = 0.05–0.3%. Linear regression analysis yielded a high R 2 value, which shows that Young’s modulus is linearly proportional to the Bis concentration within this range. Supplementary material 1 (TIFF 2931 kb)
Supplementary Fig. 2
Traction recovery. (a) Calculated tangential displacement field (Uxy) from the traction fields. (b) Calculated normal displacement field (Uz) from the traction fields. (c) Contour plot of tangential traction difference (ΔTxy = ||Txy(recovered) − Txy(input)||). (d) Contour plot of tangential traction difference (ΔTz = Tz(recovered) − Tz(input)). Supplementary material 2 (TIFF 2931 kb)
Supplementary Fig. 3
Mesh size effect (simulation). (a) Contour plot of displacement difference (ΔU = ||U(1.0-μm mesh) − U(0.5-μm mesh)||) between the 0.5 and 1.0 μm mesh sizes. (b) Contour plot of displacement difference (ΔU = ||U(2.0-μm mesh) − U(0.5-μm mesh)||) between the 0.5 and 2.0 μm mesh sizes. (c) Contour plot of traction difference (ΔT = ||T(1.0-μm mesh) − T(0.5-μm mesh)||) between the 0.5 and 1.0 μm mesh sizes. (d) Contour plot of traction difference (ΔT = ||T(2.0-μm mesh) − T(0.5-μm mesh)||) between the 0.5 and 2.0 μm mesh sizes. U = magnitude of displacement and T = magnitude of traction. Supplementary material 3 (TIFF 2931 kb)
Supplementary Fig. 4
Mesh size effect (cell data). (a) Contour plot of displacement difference (ΔU = ||U(1.0-μm mesh) − U(0.5-μm mesh)||) between the 0.5 and 1.0 μm mesh sizes. (b) Contour plot of displacement difference (ΔU = ||U(2.0-μm mesh) − U(0.5-μm mesh)||) between the 0.5 and 2.0 μm mesh sizes. (c) Contour plot of traction difference (ΔT = ||T(1.0-μm mesh) − T(0.5-μm mesh)||) between the 0.5 and 1.0 μm mesh sizes. (d) Contour plot of traction difference (ΔT = ||T(2.0-μm mesh) − T(0.5-μm mesh)||) between the 0.5 and 2.0 μm mesh sizes. U = magnitude of displacement and T = magnitude of traction. Supplementary material 4 (TIFF 2931 kb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Hur, S.S., Zhao, Y., Li, YS. et al. Live Cells Exert 3-Dimensional Traction Forces on Their Substrata. Cel. Mol. Bioeng. 2, 425–436 (2009). https://doi.org/10.1007/s12195-009-0082-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12195-009-0082-6