Abstract
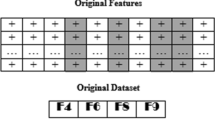
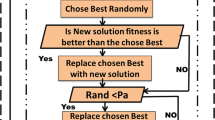
Cancer is deadly diseases still exist with a lot of subtypes which makes lot of challenges in a biomedical research. The data available of gene expression with relevant gene selection with eliminating redundant genes is challenging for role of classifiers. The availability of multiple scopes of gene expression data is curse, the selection of gene is play vital role for refining gene expression data classification performance. The major role of this article is to derive a heuristic approach to pick the highly relevant genes in gene expression data for the cancer therapy. This article demonstrates a modified bio-inspired algorithm namely cuckoo search with crossover (CSC) for choosing genes from technology of micro array that are able to classify numerous cancer sub-types with extraordinary accuracy. The experiment results are done with five benchmark cancer gene expression datasets. The results depict that CSC is outperforms than CS and other well-known approaches. It returns 99% accuracy in a classification for the dataset namely prostate, lung and lymphoma for top 200 genes. Leukemia and colon dataset CSC is 96.98% and 98.54% respectively.









Similar content being viewed by others
References
Altman NS (1995) An introduction to kernel and nearest-neighbor nonparametric regression. Am Stat 46(3):175–185
BilBan M, Buehler LK, Head S, Desoye G, Quaranta V (2002) Normalizing DNA microarray data. Mol Biol 4:57–64
Cho SB, Won HH (2003) Machine learning in DNA microarray analysis for cancer classification. In: Proc Asia-Pac Bioinf Conf Bioinf, Adelaide, Australia, pp 189–198
Ding J, Wang Q, Zhang Q, Ye Q, Ma Y (2019) A hybrid particle swarm optimization-cuckoo search algorithm and its engineering applications. Math Probl Eng. https://doi.org/10.1155/2019/5213759
Dong X, Peng Q, Wu H, Chang Z, Yue Y, Zeng Y (2019) New principle for busbar protection based on the Euclidean distance algorithm. PLoS One 14(7):e0219320
Duan KB, Rajapakse JC, Wang H, Azuaje F (2005) Multiple SVM-RFE for gene selection in cancer classification with expression data. IEEE Trans Nano Biosci 4(3):228–234
Goldberg DE (2009) Genetic algorithms in search, optimization and machine learning. Addison-Wesley, Boston
Li Y, Kang K, Krahn JM, Croutwater N, Lee K, Umbach DM, Li L (2017) A comprehensive genomic pan-cancer classification using The Cancer Genome Atlas gene expression data. BMC Genomics 18(1):1–18
Liu H, Liu L, Zhang H (2010) Ensemble gene selection for cancer classification. Pattern Recogn 43(8):2763–2772
Madhavan P, Thamizharasi V, Ranjith Kumar MV, Suresh Kumar A, Jabin MA, Sampathkumar A (2019) Numerical investigation of temperature dependent water infiltrated D-shaped dual core photonic crystal fiber (D-DC-PCF) for sensing applications. Results Phys 13:102289
Maji P (2012) Mutual information-based supervised attribute clustering for microarray sample classification. IEEE Trans Knowl Data Eng 24(1):127–140
Maulik U, Chakraborty D (2014) Fuzzy preference based feature selection and semisupervised SVM for cancer classification. IEEE Trans Nanobiosci 13(2):152–160
McLachlan G, Do KA, Ambroise C (2004) Analyzing microarray gene expression data. John Wiley & Sons Inc, Hoboken
Momiao X, Li W, Zhao J, Li J, Eric B (2001) Feature (gene) selection in gene expression-based tumor classification. J Mol Genet Metab 73(3):239–247
Motieghader H, Najafib A, Sadeghic B, Masoudi-Nejad A (2017) A hybrid gene selection algorithm for microarray cancer classification using genetic algorithm and learning automata. Inform Med Unlocked 9:246–254
Ramana TV, Pandian A, Ellammal C, Jarin T, Rashed ANZ, Sampathkumar A (2019) Numerical analysis of circularly polarized modes in coreless photonic crystal fiber. Results Phys 13:102140
Ren Z, Wang W, Li J (2016) Identifying molecular subtypes in human colon cancer using gene expression and DNA methylation microarray data. Int J Oncol 48(2):690–702
Sampathkumar A, Vivekanandan P (2018) Gene selection using multiple queen colonies in large scale machine learning. Int J Electr Eng 9(6):97–111
Sampathkumar A, Vivekanandan P (2019) Gene selection using PLOA method in microarray data for cancer classification. J Med Imaging Health Inform 9(6):1294–1300
Simon R (2009) Analysis of DNA microarray expression data. Best Pract Res Clin Haematol 22(2):271–282
Tabakhi S, Najafi A, Ranjbar R, Moradi P (2015) Gene selection for microarray data classification using a novel ant colony optimization. Neurocomputing 168:1024–1036
Tsai YS, Aguan K, Pal NR, Chung IF (2011) Identification of single-and multiple-class specific signature genes from gene expression profiles by group marker index. PLoS One 6(9):e24259
Wang X, Gotoh O (2009) Accurate molecular classification of cancer using simple rules. BMC Med Genomics 2:64
Yang XS, Deb S (2010) Cuckoo search via lévy flights. In: Proceedings of World Congress on Nature and Biologically Inspired Computing, India, IEEE Publications, USA, pp 210214
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sampathkumar, A., Rastogi, R., Arukonda, S. et al. An efficient hybrid methodology for detection of cancer-causing gene using CSC for micro array data. J Ambient Intell Human Comput 11, 4743–4751 (2020). https://doi.org/10.1007/s12652-020-01731-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12652-020-01731-7