Abstract
In recent years, there has been considerable interest and research activity in using gold nanoparticle materials for biomedical applications including biomolecular detection, bioimaging, drug delivery, and photothermal therapy. In order to apply gold nanoparticles in the real biological world, we need to have a better understanding of the potential interactions between gold nanoparticle materials and biomolecules in vivo and in vitro. Here, we report the use of dynamic light scattering (DLS) for gold nanorods characterization and nanorod–protein interaction study. In the size distribution diagram, gold nanorods with certain aspect ratios exhibit two size distribution peaks, one with an average hydrodynamic diameter at 5–7 nm, and one at 70–80 nm. The small size peak is attributed to the rotational diffusion of the nanorods instead of an actual dimension of the nanorods. When proteins are adsorbed to the gold nanorods, the average particle size of the nanorods increases and the rotational diffusion-related size distribution peak also changes dramatically. We examined the interaction between four different proteins, bovine serum albumin, human serum albumin, immunoglobulin G, and immunoglobulin A (IgA) with four gold nanorods that have the same diameter but different aspect ratios. From this study, we found that protein adsorption to gold nanorods is strongly dependent on the aspect ratio of the nanorods, and varies significantly from protein to protein. The two serum albumin proteins caused nanorod aggregation upon interaction with the nanorods, while the two immunoglobulin proteins formed a stable protein corona on the nanorod surface without causing significant nanorod aggregation. This study demonstrates that DLS is a valuable tool for nanorod characterization. It reveals information complementary to molecular spectroscopic techniques on gold nanorod–protein interactions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gold nanoparticle materials have attracted considerable attention due to their unique properties and potential applications as optical probes. Within their surface plasmon resonance (SPR) wavelength region, gold nanoparticles absorb and/or scatter light intensely, and such properties make them excellent optical materials for bioimaging, biomolecular detection, and photothermal therapy [1–4]. Gold nanoparticles can be made in a wide range of shapes and geometries such as spherical particles, nanorods, nanoshells, nanostars, and nanocages [5–10]. The optical properties of gold nanoparticles are strongly dependent on the shape and size of the particles [11–13]. For in vivo biomedical applications, gold nanoparticles with SPR band in the near-infrared (IR) region (700–900 nm) are preferred because light within this spectrum window can penetrate tissue more deeply than the visible light, and also is not substantially absorbed by the aqueous environment. Gold nanorods (GNRs) exhibit two SPR bands, the transverse band around 520–600 nm, and the longitudinal band in the near IR region, with the exact wavelength tunable by controlling the aspect ratio of the nanorods [13]. Because of their near IR SPR band, gold nanorods are considered as more promising than solid spherical nanoparticles for in vivo biomedical applications.
The potential interactions between nanoparticle materials and various biomolecules, particularly proteins, are a major research topic [14, 15]. These interactions can play a significant role on the biological activity, stability, outcome, and toxicity of the nanoparticle bioconjugate materials in vitro and in vivo. Dobrovalskaia et al. conducted a systematic study on blood plasma protein adsorption to citrate-protected gold nanoparticles and identified about 60 different proteins in the “protein corona” that is formed on the gold nanoparticle surface [16]. De Paoli Lacerda et al. reported an interaction study of common human blood proteins with spherical gold nanoparticles and determined their different binding affinities [17]. Our group recently discovered and developed a simple serum–gold nanoparticle adsorption assay for cancer detection based on the serum protein–gold nanoparticle interactions [18, 19]. We found that serum proteins adsorbed to gold nanoparticles from cancer patients differ from the normal healthy donors, and this difference can be used to predict the aggressiveness of cancer. More recently, Arvizo et al. reported a serum-adsorbed gold nanoparticle system to identify potential therapeutic targets for cancer [20].
There have been many methods and techniques reported for nanoparticle–protein interaction studies. From a literature survey, we found that the most commonly used method is fluorescence spectroscopy [16, 21, 22]. Almost all proteins have fluorescent amino acids, tryptophan, and tyrosine. When the fluorescent tryptophan or tyrosine interacts with gold nanoparticles, the fluorescence properties of tryptophan or tyrosine will change. These changes can occur as emission wavelength red-shift or blue-shift, fluorescence quenching or enhancement. Such changes have been used to determine the binding affinity and binding constant of proteins with gold nanoparticles quantitatively. However, there is a general concern on the fluorescence technique: a protein can contain multiple tryptophan and tyrosine residues. Depending on their actual distance to the gold metal core, the fluorescence of these amino acid residues can be quenched or enhanced [23, 24]. This problem could cause uncertainty in the quantitative analysis. In addition to fluorescence, circular dichroism [22, 25, 26] and FT-IR spectroscopy [22] are often used to monitor protein conformation change upon binding with gold nanoparticles. Calzolai et al. identified ubiquitin–gold nanoparticle interaction site using NMR spectroscopy [27]. All in all, multiple analytical techniques are needed to probe the complex interactions between proteins and gold nanoparticle materials.
Dynamic light scattering (DLS) is a technique that is used routinely for nanoparticle size analysis. Proteins are macromolecules. The hydrodynamic diameters of typical proteins are in the nanometer range (1–10 nm). When proteins are adsorbed to gold nanoparticles, the size of the nanoparticles will increase. We and many other groups have previously demonstrated that DLS can be used as a very convenient and powerful tool to monitor specific binding and non-specific adsorption of proteins to spherical gold nanoparticles [28–31]. Based on the nanoparticle size change, we and others have developed a novel platform technology, nanoparticle-enabled dynamic light scattering assay (NanoDLSay™) for biological and chemical detection and analysis with high to ultrahigh sensitivity and excellent reproducibility [32–38]. In this work, we applied the DLS technique to study the gold nanorods and nanorod–protein interactions. Compared to citrate-protected spherical gold nanoparticles, the understanding of nanorod–protein interactions is substantially less. Nanorod–protein interactions are more complicated than that of spherical particles, due to their non-spherical geometry and different surface ligand layer than the citrate-protected spherical nanoparticles. The goal of this study is to find better conditions for making gold nanorods–protein bioconjugates for in vivo and in vitro applications.
Experimental
Reagents
CTAB (cetyl trimethylammonium bromide)-protected Gold nanorods (CTAB-GNRs, A-12-25-550, A-12-25-600, A-12-25-650, and A-12-25-700) were purchased from Nanopartz Inc. (Loveland, Co). Spherical, citrate-protected gold nanoparticles with an average diameter of 100 nm (GNP100, catalog number 15708–9) were purchased from Ted Pella Inc. (Redding, CA). The physical properties of the four gold nanorods and nanoparticles used in this study are summarized in Table 1. The four gold nanorods are denoted as GNR1.4, GNR1.9, GNR2.4, and GNR3.0, according to their aspect ratio. All four gold nanorods have the same diameter of 25 nm. Bovine serum albumin (BSA, A7888), human serum albumin (HSA, A9511) were purchased from Sigma (Saint Louis, MO). Human immunoglobulin G (IgG) (ab91102) and IgA (ab91025) proteins were purchased from Abcam (Cambridge, MA).
UV–vis absorption spectroscopy measurement
The UV–vis absorption spectra were obtained with an Agilent 8453 spectrometer using a 1-cm path length quartz cuvette. The reference samples were deionized water.
DLS measurements
The hydrodynamic diameters of the nanoparticles under investigation were measured using a Zetasizer Nano ZS90 DLS system equipped with a red laser (532 nm) and an Avalanche photodiode detector (quantum efficiency >50 % at 532 nm; Malvern Instruments Ltd., England). A Hellma cuvette QS 3 mm was used as a sample container. DTS applications 5.10 software was used to analyze the data. All sizes reported here were based on intensity average. The intensity average particle size was obtained using a non-negative least squares analysis method. For each sample, one measurement was conducted with a fixed run time of 10 s. A detection angle of 90° was used for the size measurement.
Gold nanoparticle–protein adsorption study
To 100 μL of a GNR solution was added 2 μL of a protein solution. All protein solutions had a concentration of 1 mg/ml in 10 mM phosphate buffer. The average particle size of the mixed solution was measured after different incubation times.
Results and discussions
DLS analysis of CTAB-GNRs
We first used DLS to analyze the pure CTAB-GNRs. It should be noted that the DLS instrument used in the present study has a fixed detection angle of 90°. Particles with non-spherical shapes such as nanorods, can be more precisely characterized by multiple angles or depolarized DLS measurements [39–42]. However, because most users have fixed angle DLS instruments in their laboratories, therefore, we focused on the use of fixed angle DLS for nanorod characterization in this study.
The four GNRs have the same diameter of 25 nm, with different aspect ratios, 1.4, 1.9, 2.4, and 3.0, respectively (the four GNRs are denoted as GNR1.4, GNR1.9, GNR2.4, and GNR3.0, respectively). The longitudinal SPR peak of the four nanorods are 538, 580, 650, 698 nm, and the SPR peak of GNP100 is 560 nm (Fig. 1a). The size distribution curves of the four GNRs and GNP100 are shown in Fig. 1b. From the comparison, we noticed a number of interesting features that are absent from spherical gold nanoparticles: (1) all four nanorods showed a major particle size distribution peak at an average diameter of 75 nm; (2) with the exception of GNR1.4, other three GNRs exhibit a small particle size peak around 5–6 nm; and (3) the intensity distribution (expressed as the percentage of the total scattered light intensity) of the small size peak increases with increased aspect ratio. Figure 1c is a plot of the intensity distribution of the small size peak versus the aspect ratio of the nanorods. For comparison purposes, we also included spherical gold nanoparticles with an average diameter around 100 nm in the study. Only one major peak with an average diameter of 105 nm was observed from the spherical nanoparticle.
The size distribution peak at 75 nm is worthy for further discussion. First, this number does not correspond to either the longitudinal or the transverse dimension of the nanorods. The diameter is 25 nm for all four nanorods and the length is 34, 47, 60, and 73 nm, respectively. Second, the particle size value of this distribution peak is the same for all four nanorods, despite their obviously different mass. DLS obtains the nanoparticle size information by measuring the diffusion coefficient of the particle [43, 44]. This is the reason why the particle size obtained from DLS analysis is called the “hydrodynamic” dimension. The diffusion coefficient of a particle is not only dependent on the mass of the particle, but also the shape and the surface chemistry of the particles because these parameters affect the particle–solvent interactions, and therefore, the Brownian motion of the particles. The observed results simply mean that the four nanorods have the same diffusion coefficient as a spherical gold nanoparticle with a hydrodynamic diameter of 75 nm. In the case of nanorods, the shape and the surface chemistry of the nanorod perhaps play a more dominant role than the mass in its translational diffusion coefficient.
The small size peak around 5–7 nm is sometimes mistaken as the presence of small particle impurities. Several reports recently proposed that this small size peak is actually a representation of the rotational diffusion of the non-spherical nanorods [45, 46]. It is not an actual particle size distribution peak. It corresponds to neither the longitudinal and transverse dimension of the nanorods. This peak signifies that the rotational diffusion coefficient of the nanorods is equivalent to the translation diffusion coefficient of a spherical particle with an average diameter of 5–7 nm. The rotational diffusion appears to be strongly dependent on the aspect ratio of the nanorods: the intensity distribution (percent) of this peak increases significantly with increased aspect ratio (Fig. 1c). For GNR with an aspect ratio of 1.4, only the large particle size peak was observed, similar to the spherical nanoparticles. The findings from this study suggest that when using fixed angle DLS to analyze and interpret the size of gold nanorods, it is important to understand the true meaning of the average particle size data and size distribution peaks. The relative intensity of the size peak related to the rotational diffusion coefficient of the nanorods can provide additional size information of the nanorods.
Nanorods–protein interaction analysis
For spherical gold nanoparticles, proteins are believed to adsorb to the nanoparticle surface through a combined suite of chemical interactions including electrostatic interactions, van der Waals interactions, Au–S and Au–N bonding. Gold nanorods have a different surface chemistry from the citrate-protected spherical gold nanoparticles: the citrate ligands that protect the spherical gold nanoparticles are negatively charged, while the CTAB (cetyl trimethylammonium bromide) ligands protecting the nanorods are positively charged. It is expected that proteins interact with spherical particles and nanorods differently. Because DLS can be used to monitor the nanoparticle size change continuously, we conducted the gold nanorod–protein interaction study under kinetic conditions.
Four proteins are investigated in this study: BSA, HSA, human IgG, and human IgA. In all of these protein–nanorod interaction studies, a pure phosphate buffer (PB) solution that is used to prepare the protein solutions was used as a negative control. Figure 2 is the DLS data of the GNR1.9 (A and B) and GNR2.4 (C and D) upon mixing with BSA or PB control solution. Three nanorods, GNR1.4, GNR1.9, and GNR3.0 showed no size and size distribution change at the presence of BSA (the data of GNR1.4 and GNR3.0 is not shown here, but very similar to GNR1.9). That is to say, the average particle size of the two peaks, 5–6 and 75 nm, remains unchanged (Fig. 2a) compared to phosphate buffer control solution, and the relative intensity of these two peaks remain unchanged as well (Fig. 2b), suggesting that there is no interaction between BSA and these three GNRs. Interestingly, there is a dramatic response from GNR2.4: First, the average particle size of the large size peak increases steadily and quickly from about 75 nm to more than 200 nm within 10 min of incubation time, and the average particle size of the small size peak also increases from 5–7 nm to about 20–30 nm (Fig. 2c). Second, the relative intensity of the small particle size peak decreases from the original 50 % to less than 10 % (Fig. 2d), and the relative intensity of the large particle size peak increases from 50 % to more than 90 %.
Particle size analysis results of GNR1.9 (a and b) and GNR2.4 (c and d) upon incubation with bovine serum albumin (BSA) or 10 mM PB solution (control). All samples were prepared by adding 2 μL of 1 mg/mL protein solution or PB control solution into 100 μL GNR or GNP solution. All data points are average values of three experiments. a and c are the plots of the average particle size of the mixed sample solution versus the incubation time. b and d are the plots of the peak intensity distribution (percent) versus the incubation time. Legends for all four figures are the same as a
The interaction between HSA and nanorods are very similar (Fig. 3a, b: GNR1.9; Fig. 3c, d: GNR2.4). Again, three nanorods, GNR1.4, GNR1.9, and GNR3.0, showed no response to HSA at all (only the data for GNR1.9 is presented here), while GNR2.4 exhibited substantial particle size increase upon interaction with HSA. However, there are some slight differences between HSA and BSA: the particle size increase caused by HSA appears to be even larger than that is caused by BSA, and the small size peak that is indicative of rod-shaped particle disappeared completely over incubation time.
Particle size analysis results of GNR1.9 (a and b) and GNR2.4 (c and d) upon incubation with human serum albumin (HSA) or 10 mM PB solution (control). All samples were prepared by adding 2 μL of 1 mg/mL protein solution or PB control solution into 100 μL GNR or GNP solution. All data points are average values of three experiments. a and c are the plots of the average particle size of the mixed sample solution versus the incubation time. b and d are the plots of the peak intensity distribution (percent) versus the incubation time. Legends for all four figures are the same as a
IgA and IgG are immunoglobulin proteins. IgA is a dimer of IgG, linked together through the Fc region, with the Fab region exposed outwards. Again, we did not observe any size change from GNR1.4, GNR1.9, and GNR3.0 (data not shown here). Only GNR2.4 showed response to IgA and IgG, however, in a very different way from the two serum albumin proteins (Fig. 4). The interaction of IgA or IgG caused the large particle size peak increase from 75 nm to about 100 nm for IgA (Fig. 4a), and to 85 nm for IgG (Fig. 4b). So the net increase is 15 nm for IgA and 10 nm for IgG. The small size peak, increased from about 5 to 15 nm for IgA (Fig. 4c), and from 5 nm to about 9 nm for IgG (Fig. 4d). Most noticeable is that the intensity distribution of the small size peak changed only very slightly: it decreased from 50 % to about 45–47 % (Fig. 4e, f) for both IgA and IgG.
Particle size analysis results of GNR2.4 upon incubation with immunoglobulin A (IgA) or immunoglobulin G (IgG) solution. All samples were prepared by adding 2 μL of 1 mg/mL protein solution or PB control solution into 100 μL GNR solution. a and b are the plots of the average particle size of the large size peak versus incubation time; c and d are the plots of the average particle size of the small size peak versus incubation time. e and f are the plots of the small peak intensity distribution (percent) versus incubation time
As a comparison to GNRs, the interaction between a spherical gold nanoparticle, GNP100 with a hydrodynamic diameter of 105 nm was also studied. Upon mixing with BSA and HSA at a concentration of 1 mg/mL, no obvious nanoparticle size increase was observed. Upon mixing with IgA and IgG, the nanoparticle size increased by about 20 nm, from 105 to 125 nm.
The particle size analysis revealed several unexpected results regarding the nanorod–protein interactions. The first noticeable result is that among the four nanorods studied here, only nanorods with an aspect ratio of 2.4, GNR2.4, appears to interact with the two serum albumins and the two immunoglobulins. We have not noticed similar findings from previous reports on such a phenomenon. In one study reported by Pan et al., it was mentioned that BSA adsorbs more strongly to gold nanorods with an aspect ratio of 8.0 than nanorods with an aspect ratio of 3.0 [21]. The surface chemistry of the four nanorods investigated in this study is the same, because they are all CTAB-protected, and positively charged on the surface. At this moment, the exact reason behind this difference is unclear. Since only the aspect ratio is different, we can only hypothesize that this may be related to the different stability of the CTAB bilayer structure on the nanorods. It is possible that GNR with an aspect ratio of 2.4 may have the lowest stability, therefore, more easily displaced by proteins.
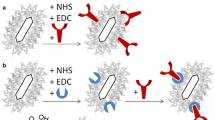
A second noticeable result is that it appears that the two serum albumin proteins (BSA and HSA) caused nanorod aggregation (Fig. 5a) while the two immunoglobulin proteins formed a stable protein corona on the nanorod surface without causing nanorod aggregation (Fig. 5b). Serum albumin is a protein with a molecular weight around 60 KDa. The hydrodynamic diameter of a serum albumin is around 5 nm. If a complete layer of BSA molecule is adsorbed to the nanoparticle, the net increase of the particle size should not exceed about 10 nm. The dramatic size increase from 75 to more than 200 nm can only be interpreted as nanorod aggregate formation. Furthermore, the aggregate formation is most likely a random aggregate. With its non-spherical geometry, GNRs are known to form end-to-end or side-by-side assemblies under various conditions [38]. From the analysis of pure GNRs, we know that with higher aspect ratio, the intensity of the small size peak increases. If BSA interacts with GNR primarily through end-to-end mode, we should see intensity increase of the small particle size peak, not decrease. As random aggregates are formed, the aggregates “look” more like a spherical particle, and the signature peak indicative of the rotational diffusion of a rod-like particle will disappear.
Schematic illustrations of two different interaction models between GNRs and proteins. Serum albumins (BSA and HSA) caused gold nanorods aggregate formation (a), while IgG and IgA formed a stable protein corona on the nanorod surface (b). Also in a, some CTAB ligands are intentionally eliminated to illustrate the potential disruption of the CTAB double-layer structure
On the other hand, the interaction of the two immunoglobulin proteins (IgG and IgA) with the nanorod GNR2.4 appears to lead to the formation of a stable protein corona on the nanorod surface. The average particle size of the nanorods increased only about 10–15 nm, which is on par with the hydrodynamic diameter of the two proteins. The relative intensity of the small particle size peak, signature of the nanorod rotational diffusion, remains almost the same. This suggests that the nanorods, after their interaction with the two immunoglobulin proteins, maintained their rod-like shape.
Currently, we do not know the exact reasons behind these differences. We offer the following explanations as possible mechanisms. The isoelectric point of serum albumin is around 4.7; and the isoelectric points of immunoglobulins are higher, typically in the range of 6.1–8.5 (refer to the product information, Sigma-Aldrich). At neutral PB buffer solution (pH 7.4), serum albumins are negatively charged. The CTAB-GNRs are positive charged. It could be that the electrostatic interaction between negatively charged serum albumin and the nanorods caused the disruption of the CTAB biolayer. GNR2.4 may have the weakest CTAB bilayer structure and is easily perturbed by protein adsorption interaction. The CTAB bilayer structure may be disrupted too quickly to allow the serum albumin proteins to form a stable protecting layer on the nanorod surface. Subsequently, the exposed nanorods quickly aggregated together due to strong van der Waals interactions between the exposed metal cores. As to the immunoglobulin proteins, these proteins are membrane proteins that are produced by the B cells and located in the membrane of B cells. The immunoglobulins may bind to the gold nanorods by first inserting the Fc region of the antibody into the CTAB bilayer, and then further interaction with the gold nanorods. The surface protection layer of the nanorods is not disrupted during the protein adsorption process. In the end, a stable protein corona is formed on the nanorods. These hypotheses certainly require further extensive studies.
Conclusion
In this study, we demonstrate that fixed angle DLS may also be used for gold nanorods characterization and study. However, one should be aware that when using a fixed angle DLS instrument as most users have, the particle size data of nanorods obtained from DLS does not represent the true physical dimension of the nanorods. The actual information determined by DLS is the diffusion coefficient of the particle. If the nanoparticle is spherical, the hydrodynamic diameter can be revealed by Stokes–Einstein equation. If the particle has a rod shape, the diffusion coefficient determined by DLS is still accurate, but the hydrodynamic diameter cannot be deduced from the Stokes–Einstein equation. Despite this fact, we can still use single, fixed angle DLS to monitor the nanorod size change upon protein interaction, as demonstrated in this study. The small particle size peak that is a signature of the rotational diffusion of the nanorods is important for determining the nanorod aggregate formation. Complementary to various molecular spectroscopic techniques, DLS can provide additional information on the complex protein–nanorod interactions. Although many previous studies have demonstrated the use of antibody-conjugated gold nanorods for imaging and biomolecular assay development [47–51], it is also acknowledged by these previous studies that there are more difficulties involved in the preparation of gold nanorod–protein bioconjugates than the spherical nanoparticle bioconjugates. More often than not, the CTAB-protected gold nanorods need to be first modified with other ligands or functional groups to facilitate bioconjugation. Our study suggests that CTAB-protected gold nanorods with certain aspect ratios can be directly conjugated to antibodies through a simple adsorption process. The aspect ratio of the GNRs should be considered in selecting the most suitable nanorods for future application development
References
Jans H, Huo Q (2012) Gold nanoparticle-enabled biological and chemical detection and analysis. Chem Soc Rev 41:2849–2866
Dykman L, Khlebtsov N (2012) Gold nanoparticles in biomedical applications: recent advances and perspectives. Chem Soc Rev 41:2256–2282
Dreaden EC, Alkilany AK, Huang X, Murphy CJ, El-Sayed MA (2012) The golden age: gold nanoparticles for biomedicine. Chem Soc Rev 41:2740–2779
Boisselier E, Astruc D (2009) Gold nanoparticles in nanomedicine: preparations, imaging, diagnostics, therapies and toxicity. Chem Soc Rev 38:1759–1782
Newhouse RJ, Zhang JZ (2012) Optical properties and applications of shape-controlled metal nanostructures. Rev Plasmonics 2010:205–238
Chen J, Saeki F, Wiley BJ, Cang H, Cobb MJ, Li Z-Y, Au L, Zhang H, Kimmey MB, Li X, Xia Y (2005) Gold nanocages: bioconjugation and their potential use as optical imaging contrast agents. Nano Lett 5(3):473–477
Hu M, Chen J, Li Z, Au L, Hartland GV, Li X, Marquez M, Xia Y (2006) Gold nanostructures: engineering their plasmonic properties for biomedical applications. Chem Soc Rev 35:1084–1094
Sun Y, Xia Y (2003) Gold and silver nanoparticles: a class of chromophores with colors tunable in the range from 400 to 750 nm. Analyst 128:686–691
Halas NJ, Lal S, Chang W, Link S, Nordlander P (2011) Plasmons in strongly coupled nanostructures. Chem Rev 111:3913–3963
Wang H, Brandl D, Nordlander P, Halas NJ (2006) Plamonic nanostructures: artificial molecules. Acc Chem Res 40:53–62
Jain PK, Lee KS, El-Sayed IH, El-Sayed MA (2006) Calculated absorption and scattering properties of gold nanoparticles of different size, shape, and composition: applications in biological imaging and biomedicine. J Phys Chem B 110:7238–7248
Nehl CL, Liao H, Hafner JH (2006) Optical properties of star-shaped gold nanoparticles. Nano Lett 6:683–688
Link S, Mohamed MB, El-Sayed MA (1999) Simulation of the optical absorption spectra of gold nanorods as a function of their aspect ratio and the effect of the medium dielectric constant. J Phys Chem B 103:3073–3077
Mout R, Moyano DF, Rana S, Rotello VM (2012) Surface functionalization of nanoparticles for nanomedicine. Chem Soc Rev 41:2539–2544
Walkey CD, Chan WCW (2012) Understanding and controlling the interaction of nanomaterials with proteins in a physiological environment. Chem Soc Rev 41:2780–2799
Dobrovolskaia MA, Patri AK, Zheng J, Clogston JD, Ayub N, Aggarwal P, Neun BW (2009) Interaction of colloidal gold nanoparticles with human blood: effects on particle size and analysis of plasma protein binding profiles. Nanomedicine (Nanotechnol Biol Med) 5:106–117
Lacerda SHDP, Park JJ, Meuse C, Pristinski D, Becker ML, Karim A, Douglas JF (2010) Interaction of gold nanoparticles with common human blood proteins. ACS Nano 4:365–379
Huo Q, Colon J, Codero A, Bogdanovic J, Baker CH, Goodison S, Pensky MY (2011) A facile nanoparticle immunoassay for cancer biomarker discovery. J Nanobiotechnol 9:20, open access
Huo Q, Litherland SA, Sullivan S, Hallquist H, Decker DA, Rivera-Ramirez I (2012) Developing a nanoparticle test for prostate cancer scoring. J Transl Med 10:44, open access
Arvizo RR, Giri K, Moyano D, Miranda OR, Madden B, McCormick DJ, Bhattacharya R, Rotello VM, Kocher JP, Mukherjee P (2012) Identifying new therapeutic targets via modulation of protein corona formation by engineered nanoparticles. PLoS One 7:e33650, open access
Pan B, Cui D, Xu P, Li Q, Huang T, He R, Gao F (2007) Study on interaction between gold nanorod and bovine serum albumin. Colloids Surf A 295:217–222
Shang L, Wang Y, Jiang J, Dong S (2007) pH-dependent protein conformational changes in albumin:gold nanoparticle bioconjugates: a spectroscopic study. Langmuir 23:2714–2721
Iosin M, Toderas F, Baldeck PL, Astilean S (2009) Study of protein-gold nanoparticle conjugates by fluorescence and surface-enhanced Raman scattering. J Mol Struct 924–926:196–200
Matveeva EG, Shtoyko T, Gryczynski I, Zkopova I, Gryczynski Z (2009) Fluorescence quenching/ enhancement surface assays: signal manipulation using silver-coated gold nanoparticles. Chem Phys Let 454:85–90
Fischer NO, McIntosh CM, Simard JM, Rotello VM (2002) Inhibition of chymotrypsin through surface binding using nanoparticle-based receptor. Proc Natl Acad Sci 99:5018–5023
Hong R, Fischer NO, Verma A, Goodman CM, Emrick T, Rotello VM (2004) Control of protein structure and function through surface recognition by tailored nanoparticle scaffolds. J Am Chem Soc 126:739–743
Calzolai L, Franchini F, Gilliland D, Rossi F (2010) Protein–nanoparticle interaction: identification of the ubiquitin–gold nanoparticle interaction site. Nano Lett 10:3101–3105
Jans H, Liu X, Austin L, Maes G, Huo Q (2009) Dynamic light scattering as a powerful tool for gold nanoparticle bioconjugation and biomolecular binding study. Anal Chem 81:9425–9432
Austin L, Liu X, Huo Q (2010) An immunoassay for monoclonal antibody isotyping and quality analysis using gold nanoparticles and dynamic light scattering. Am Biotechnol Lab 28(8):10–12
Khlebtsov NG, Bogatyrev VA, Khlebtsov BN, Dykman LA, Englebienne P (2003) A multilayer model for gold nanoparticle bioconjugates: application to study of gelatin and human IgG adsorption using extinction and light scattering spectra and the dynamic light scattering method. Colloid J 65:679–693
Tsai DH, Delrio FW, Keene AM, Tyner KM, MacCuspie RI, Cho TJ, Zachariah MR, Hackley VA (2011) Adsorption and conformation of serum albumin protein on gold nanoparticles investigated using dimensional measurements and in situ spectroscopic methods. Langmuir 27:2464–2477
Liu X, Dai Q, Austin L, Coutts J, Knowles G, Zou J, Chen H, Huo Q (2008) A one-step homogeneous immunoassay for cancer biomarker detection using gold nanoparticle probes coupled with dynamic light scattering. J Am Chem Soc 130:2780–2782
Dai Q, Liu X, Coutts J, Austin L, Huo Q (2008) A one-step highly sensitive method for DNA detection using dynamic light scattering. J Am Chem Soc 130:8138–8139
Kalluri JR, Arbneshi T, Khan SA, Nelly A, Candice P, Varisli B, Washington M, McAfee S, Robinson B, Banerjee S, Singh AK, Senapati D, Ray PC (2009) Use of gold nanoparticles in a simple colorimetric and ultrasensitive dynamic light scattering assay: selective detection of arsenic in groundwater. Angew Chem Int Ed 48:9668–9671
Gao D, Sheng Z, Han H (2011) An ultrasensitive method for the detection of gene fragment from transgenics using label-free gold nanoparticle probe and dynamic light scattering. Anal Chim Acta 696:1–5
Driskell JD, Jones CA, Tompkins SM, Tripp RA (2011) One-step assay for detecting influenza virus using dynamic light scattering and gold nanoparticles. Analyst 136:3083–3090
Wang X, Ramström O, Yan M (2011) Dynamic light scattering as an efficient tool to study glyconanoparticle-lectin interactions. Analyst 136:4174–4178
Wang L, Zhu Y, Xu L, Chen W, Kuang H, Liu L, Agarwal A, Xu C, Kotov NA (2010) Side-by-side and end-to-end gold nanorod assemblies for environmental toxin sensing. Angew Chem Int Ed 49:5472–5475
Pecora R (1968) Spectrum of light scattered from optically anisotropic macromolecules. J Chem Phys 49:1036–1043
Zero K, Pecora R (1985) Dynamic depolarized light scattering. In: Pecora R (ed) Dynamic light scattering applications of photon correlation spectroscopy. Plenum, New York, pp 83–99
Van der Zande BMI, Dhont JKG, Bohmer MR, Philipse AP (2000) Collidal dispersions of gold rods characterized by dynamic light scattering and electrophoresis. Langmuir 16:459–464
Lehner D, Lindner H, Glatter O (2000) Determination of the translational and rotational diffusion coefficients of rodlike particles using depolarized dynamic light scattering. Langmuir 16:1689–1695
Berne BJ, Pecora R (1976) Dynamic light scattering: with applications to chemistry, biology, and physics. Wiley, New York
A Technical Note from Malvern Instruments: http://www.malvern.com/common/downloads/campaign/MRK656-01.pdf
Rodrígues-Fernández J, Pérez-Juste J, Liz-Marzán LM, Lang PR (2007) Dynamic light scattering of short Au rods with low aspect ratios. J Phys Chem C 111:5020–5025
Khlebtsov BN, Khlebtsov NG (2011) On the measurement of gold nanoparticle sizes by the dynamic light scattering method. Colloid J 73:118–127
Wang C, Chen Y, Wang T, Ma Z, Su Z (2007) Biorecognition-driven self-assembly of gold nanorods: a rapid and sensitive approach toward antibody sensing. Chem Mater 19:5809–5811
Tong L, Wei Q, Wei A, Cheng J (2009) Gold nanorods as contrast agents for biological imaging: optical properties, surface conjugation and photothermal effects. Photochem Photobiol 85:21–32
Pissuwan D, Valenzuela SM, Killingsworth MC, Xu X, Cortie MB (2007) Targeted destruction of murine macrophase cells with bioconjugated gold nanorods. J Nanoparticle Res 9:1109–1124
Pissuwan D, Valenzuela SM, Miller CM, Cortie MB (2007) A golden bullet? Selective targeting of Toxoplasma gondii tachyzoites using antibody-functionalized gold nanorods. Nano Lett 7:3808–3812
Yu C, Irudayaraj J (2007) Multiplex biosensor using gold nanorods. Anal Chem 79:572–579
Acknowledgment
This work is supported by a State of Florida Boost Scholar Award to Q.H. The authors also want to thank the reviewers of this manuscript for their highly constructive comments and suggestions, which have led to significant improvements of the paper.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Helin Liu and Nickisha Pierre-Pierre made equal contributions to this work.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Liu, H., Pierre-Pierre, N. & Huo, Q. Dynamic light scattering for gold nanorod size characterization and study of nanorod–protein interactions. Gold Bull 45, 187–195 (2012). https://doi.org/10.1007/s13404-012-0067-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13404-012-0067-4