Abstract
In this work, three new water-soluble aluminum(III), gallium(III) and indium(III) Schiff base complexes; [M(L)NO3], where L denotes; N,N′-bis(5-sulfosalicyliden)-1,2-phenylendiamin (5-SO3-1,2-salophen); were synthesized and characterized by UV–Vis spectroscopy, 1H NMR, FT-IR, thermal gravimetry and C.H.N analyses. Also, the optimized structure and the energy level of the frontier orbitals for the complexes were computed using B3LYP/LANL2DZ method and 6–311g basis set. The interaction of these complexes with bovine serum albumin (BSA) was studied by fluorescence and UV–Vis techniques. The results showed that the intrinsic fluorescence of BSA was quenched by adding the complexes, which was rationalized in terms of the dynamic quenching mechanism. Binding constants, Stern–Volmer constants, number of binding sites and binding stoichiometry were determined by fluorescence method. Furthermore, the thermodynamic parameters calculating by van’t Hoff equation, indicated that the binding mode is entropy driven and enthalpically disfavored. Site marker competitive experiments revealed that these complexes can bind to the subdomain IIIA (site II) of BSA. Based on the Förster theory of non-radiation energy transfer, the efficiency of energy transfer and the distance between the donor (BSA) and the acceptor (complex) were obtained. Also, the synchronous fluorescence spectra displayed that the microenvironment of the tyrosine residues was not apparently changed. Finally, the binding of the complexes to BSA was modeled by molecular docking method. Excellent agreement was found between the experimental and theoretical results with respect to the mechanism of binding and binding constants. Results indicated that the order of propensity of the complexes to BSA was as follows: [In(L)NO3] > [Ga(L)NO3] > [Al(L)NO3].
Similar content being viewed by others
Introduction
Schiff base complexes are important class of metal complexes in medicinal and pharmaceutical fields. In recent years, they have shown biological applications including excellent antibacterial, antifungal [1] and anticancer properties [2]. Diamino-tetradentate Schiff base ligands and their complexes have been used as biological models in understanding the structure of biomolecules and biological processes. Metal complexes containing N2O2 Schiff base ligands have been attracted some attention in biological systems [3]. Most of the studies have been done on the transition metal complexes, but main group metals especially group 13 metals were not considered widely [4–7]. In recent years, the role played by aluminum in causing Alzheimer’s disease has been clear. The accumulation of aluminum in the brain causes Alzheimer’s disease [8]. Thus, aluminum and its complexes (such as Schiff base complexes) and their biological activity attract some attentions [9]. Other group 13 Schiff base complexes [gallium(III) and indium(III)] [10] are proven to have antimalaria properties [11–13], however, Ga(III) Schiff base complexes were previously reported as potential positron emitting (PET) radiopharmaceuticals with utility as myocardial perfusion imaging agents [14].
Serum albumins are the most abundant proteins in the plasma. The most important property of this group of proteins is that they serve as a depot protein and as a transport protein for many drugs and other bioactive small molecules [15]. Many drugs and bioactive molecules bind to albumin as carrier. Then, it is necessary and important to study the interaction of synthetic compounds as drug with these proteins [16]. Albumins from blood plasma bind with pyridoxal phosphate, cysteine, glutathione, Schiff base ligands [17], various metal ions such as Cu(II), Ni(II), Mn(II), Co(II), Hg(II), Ga(III), metal complexes [18] and metallothionein(a metal-binding protein) [19, 20].
Most ligands that are specifically designed for water solubility contain hydrophilic groups such as phosphonium, sulfonate and carboxylate moieties to increase the solubility of these compounds in water [21].
Fluorescence spectroscopy is a powerful method for the study of the interaction of chemical and biological systems since it allows non-intrusive measurements of substances in low concentration under physiological conditions. This technique is one of the most important methods to study the interaction between drugs and proteins. The spectral changes observed on the binding of fluorophores with proteins are an important tool for the investigations of the topology of binding sites, conformational changes and characterization of substrate to ligand binding. Here, the quenching of the intrinsic tryptophan fluorescence of BSA used as a tool to study the interaction of the metal complexes with this transport protein under physiological condition [22].
In continuing our research on the interaction of transition metal complexes with BSA and HSA, in this study, some new water-soluble group 13 metals [Al(III), Ga(III) and In(III)] Schiff base complexes have been synthesized to study the effect of metal on the mechanism of binding and binding constants of the complexes with bovine serum albumin (BSA). Both experimental and theoretical results indicated that indium(III) complex more strongly interacted with BSA than gallium(III) and aluminum(III). These studies are useful for the design and synthesis of new metal-based drugs.
Experimental
Materials and instruments
All chemicals were used as obtained commercially, without further purification. The 1H NMR spectra were recorded by Bruker Avance DPX 250MHz spectrometer. UV–Vis measurements were carried out in Perkin-Elmer (LAMBDA 2) UV–Vis spectrophotometers. FT-IR spectra were recorded by Shimadzu FT-IR 8300 infrared spectrophotometer. Elemental analysis was carried out by Thermo Fininngan-Flash 1200. Thermogravimetric measurements were performed on a Perkin-Elmer Pyris Diamond Model. Melting points were determined on a BUCHI 535. Fluorescence spectra were carried out on a Perkin-Elmer (LS-45) spectrofluorimeter equipped with a thermostat bath at different temperatures. All experiments were carried out in triple distilled water at pH = 7.4, 100 mM Tris–HCl buffer and 50 mM NaCl.
Synthesis and characterization
Sodium salicylaldehyde-5-sulfonate monohydrate (sals) was synthesized according to the literature procedure [16] and the sulfonation was carried out at 100 °C.
Synthesis of N,N′-bis(5-sulfosalicyliden)-1,2-phenylendiamin (H2L)
1,2-Phenylendiamin(1.0 mmol) in water (20 mL) was added dropwise with continuous stirring to a warm ethanolic solution of sals (2.0 mmol). The mixture was refluxed for 5 h. During this time, a yellow precipitate was formed then filtered and washed with cool ether. The resulting yellow compound was dried in vacuum at 50 °C.
H2L
Yield: 38 % m.p. >250 °C, color: yellow, 1H NMR (250 MHz, DMSO-d6, δ ppm): 13.19 (s, 2H, OH), 8.97 (s, 2H, N=CH), 6.86–7.94 (m, 10H, Ar–H). FT-IR (KBr, cm−1): 3386 ν (O–H), 1620 ν (C=N), 1481 ν (C=C), 1033, 1110, 1195 \(\nu_{{({\text{SO}}_{3}^{ - } )}}\). Anal. Found (Calc.) %: C20H14N2S2O8Na2, C: 46.77 (46.34); H: 2.19 (2.33); N: 5.38 (5.40); UV–Vis (H2O) λ (nm): 325, 259, 225.
Synthesis of the complexes
A solution of metal(III) nitrate (2 mmol was dissolved in minimum amount of water) was added to the stirring solution of the ligand (H2L) (2 mmol). The mixture was vigorously stirred for 6 h at 40 °C, and then ethanol (5 mL) was added and stirring was continued to precipitate the product. The resulting powder was collected by filtration, washed with ethanol and ether then recrystallized from ethanol and dried at 50 °C in vacuum (Scheme 1).
Sodium [(N,N′-bis(5-sulfosalicyliden)-1,2-phenylendiamino)nitrato]aluminate(III) Na2 [Al(5-SO3-1,2-salsophen)NO3]
Yield: 47 %. m.p. >250 °C, color: yellow.1H NMR (250 MHz, DMSO-d6, δ ppm): 8.38 (s, 2H, N=CH), 7.08–7.76 (m, 10H, Ar–H). FT-IR (KBr, cm−1): 3425 ν (O–H), 1610 ν (C=N), 1465 ν (C=C), 1388, 820 ν (N–O), 1026, 1110, 1180 \(\nu_{{({\text{SO}}_{ 3} )}}\), 563 ν (M–N), 486 ν (M–O). Anal. Found (Calc.) %: (C20H12AlN3Na2O11S2); C: 39.79 (39.55); H: 1.93 (1.99); N: 6.66 (6.92); UV–Vis (H2O) λ (nm): 360, 250, 230.
Sodium [N,N′-bis(5-sulfosalicyliden)-1,2-phenylendiamino)nitrato]galliumate(III) Na2 [Ga(5-SO3-1,2-salsophen)NO3]
Yield: 46 %. m.p. >250 °C, color: yellow, 1H NMR (250 MHz, DMSO-d6, δ ppm): 9.46 (s, 2H, N=CH), 6.89–8.22 (m, 10H, Ar–H). FT-IR (KBr, cm−1): 3425 ν (O–H), 2950–3075 ν (C–H), 1612 ν (C=N), 1458 ν (C=C), 1382, 825 ν (N–O), 1026, 1110, 1188 \(\nu_{{({\text{SO}}_{ 3} )}}\), 543 ν (M–N), 455 ν (M–O). Anal. Found (Calc.) %:(C20H12GaN3Na2O11S2); C: 36.81 (36.95); H: 1.71 (1.86); N: 6.80 (6.46); UV–Vis (H2O) λ (nm): 370, 250, 235.
Sodium [(N,N′-bis(5-sulfosalicyliden)-1,2-phenylendiamino)nitrato]indiumate(III) Na2[In(5-SO3-1,2-salsophen)NO3]
Yield: 58 %. m.p. >250 °C, color: yellow, 1H NMR (250 MHz, DMSO-d6, δ ppm): 9.39 (s, 1H, N=CH), 6.84–8.22 (m, 10H, Ar–H). FT-IR (KBr, cm−1): 3445 ν (O–H), 2950–3080 ν (C–H), 1612 ν (C=N), 1460 ν (C=C), 1458, 1380, 1320, 1020, 815 ν (N=O), 1378 ν (N–O), 1181, 1110, 1028 \(\nu_{{({\text{SO}}_{ 3} )}}\), 552 ν (M–N), 480 ν (M–O). Anal. Found (Calc.) %: (C20H12InN3Na2O11S2); C: 34.79 (34.55); H: 1.93 (1.74); N: 5.86 (6.04); UV–Vis (H2O) λ (nm): 370, 250, 225.
Methods
Absorption spectroscopy
Absorption titration was performed using Perkin-Elmer (LAMBDA 2) UV–Vis spectrophotometer, equipped with a thermostat bath. Temperature kept constant within ±0.1 °C. Absorption titrations were performed at 280 nm at fixed concentration of BSA (1 × 10−5 M), while varying the concentration of Schiff base complex. A change in absorbance at this wavelength is a convincing evidence for the formation of ligand–BSA complex.
Fluorescence titration of BSA with the complexes
2.5 mL of BSA solution (1 × 10−7 M) was poured into the fluorescence cell. Emission spectra were recorded after each addition (10 µL) of complex solutions (1.25 × 10−5 M) in the same buffer at different temperatures (298, 304, and 310 K). The samples were excited at 280 nm. The observed fluorescence intensities were also correlated for dilution.
The reaction time has been studied and the results showed that 2 min was enough for the stabilization. So the change in fluorescence emission intensity was measured within 2 min after each addition of the complex.
Competition reactions between phenylbutazone, ibuprofen and complexes for binding to BSA
To identify the binding sites of BSA, displacement of phenylbutazone and ibuprofen was followed fluorimetrically. For this purpose, an equimolar mixture of BSA 2.5 mL (1 × 10−7 M) and phenylbutazone (5 μL, 5.0 × 10−5 M) in 50:50 V/V % water/acetone solution was prepared and kept for 1 h and then titrated with complex (1.25 × 10−3 M) solution. Similar solution was prepared that containing 2.5 mL BSA (1 × 10−7 M) and 5 μL ibuprofen (5 × 10−5 M in 50:50 V/V % water/acetone) was titrated by complex (1.25 × 10−5 M) solutions.
Synchronous fluorescence
The synchronous fluorescence spectra were achieved by scanning simultaneously the excitation and emission monochromator by Perkin-Elmer (LS-45) spectrofluorimeter. The synchronous fluorescence spectra only showed the tyrosine residue and the tryptophan residue of BSA when the wavelength interval (∆λ) is 15 and 60 nm, respectively [23].
Computational details
The starting point for geometry optimization was modeled with the Gaussian View program, while geometry calculations were made using the Gaussian 03 set of programs. Calculations were performed with B3LYP method as implemented in the Gaussian programs [24]. The geometries of these complexes were fully optimized using 6-311 g basis set for all atoms and LANL2DZ basis set were performed for In(III) complex.
Molecular docking
The crystal structure of BSA (entrycodes 3V03) was taken from the Brook haven Protein Data Bank (http://www.pdb.org). 3V03 was chosen because of no missing at atoms, no crystallized ligand and having reasonable good resolution. To get the most stable geometry of Schiff base complexes, the structure optimizing calculation was carried out at the 6–311g level, by employing the Becke three-parameter Lee–Yang–Parr (B3LYP) hybrid density functional theory using the quantum chemistry software Gaussian 03. The structures with the lowest energy were selected for the docking study. All flexible ligand docking calculations were performed with the MVD program package. Bonding site was chosen by (x = 37.81, y = 19.87, z = 58.18, and radius (A) = 13) for BSA to allow the ligands to rotate freely, even in their most fully extended conformations. The molecular docking program Molegro Virtual Docker (MVD) was employed to generate a docked conformation of the complexes with BSA [25].
Results and discussion
Characterization of the complexes
FT-IR spectra
FT-IR spectrum of the ligand exhibited broad medium intensity band at 3386 cm−1 which were assigned to the phenolic O–H vibration. This band was disappeared via complexation to the metal ions. The FT-IR spectra of complexes exhibited absorption bands at around 3430 cm−1 that was attributed to the presence of the hydrated water. Azomethine vibration of the free ligand was observed around 1620 cm−1. In the complexes, these bands were shifted to the lower frequencies, which suggested the coordination of the ligand to the metal ions through the azomethine nitrogen.
A broad peak in the range 1380–1388 cm−1 and a medium peak in the range 815–825 cm−1 were assigned to the vibration of N‒O bond of NO3 − ligand. It has been found that the nitrato ligand in [InL(NO3)] complex was coordinated bidentately. In the FT-IR spectra of the complexes, the (N=O) (υ 1) and υ asym(NO2) (υ 5) vibrations characteristic of the chelating bidentate nitrate ion appear at ca. 1458 and 1320 cm−1, respectively. The υ sym(NO2) vibration (υ 2) at ca. 1026 cm−1 was characteristic of bidentate chelating nitrate. The frequency separation of the nitrate stretching fundamentals (υ 1–υ 5) has been used as a criterion to distinguish between mono- and bidentate chelating nitrates. The magnitude of this separation (Δυ 148 cm−1 for [In(L)NO3]) is indicative of bidentate coordination of the nitrate ion [26]. The M–O and M–N stretching vibrations usually appeared in the region 400–500 and 410–580 cm−1, respectively. The other series of weak bands between 3100 and 2800 cm−1 were related to C–H modes of vibrations. The ring skeletal vibrations C=C were considered in the region 1440–1481 cm−1 in all the compounds [27].
UV–Vis spectra
The spectrum of the ligand exhibited one band at 225 nm that was attributed to π → π* transitions in aromatic rings and two bands at 259 and 325 nm were assigned to π → π* and n → π* transition, respectively, which were attributed to C=N, C=O or S=O groups. The complexes showed intense absorptions at 220–370 nm, attributed to the π → π* transition in the aromatic rings or azomethine groups.
1H NMR spectra
The 1H NMR spectral data of the ligand showed singlet signal for the azomethine protons at 8.97 ppm and singlet signals for hydroxyl protons at 13.19 ppm. The signals of the hydrogens of the aromatic rings were appeared at 6.86–7.94 ppm. Peak due to OH of the Schiff base was not observed after formation of the complex [28].
Thermal analysis
In the thermal investigation, heating rate was suitably controlled at 10 °C min−1 under nitrogen atmosphere, and the weight loss was measured from the ambient temperature up to 1000 °C. The TG/DTA curves, revealed that thermal decomposition of [Al(L)NO3] occurred in one step but for [Ga(L)NO3] and [In(L)NO3] decomposition occurred in two steps. For indium complex, first mass loss (Calc. 10 %, found 9.5 %) could be seen at 180 °C and was attributed to the elimination of the NO3 group. The second mass loss (Calc. 73 %, found 75 %) was observed at 577 °C and was attributed to elimination of organic part of the complex (C27H16N2O8). The results of TGA data for all complexes are summarized in Table 1.
Geometry optimization and energies of the complexes
The optimized stable molecular structures of the complexes are shown in Fig. 1. Selected bond lengths and bond angles of the compounds are listed in Table 2. To check the validity of the method applied, we used the X-ray diffraction data of a similar complex to compare the optimized structures of the synthesized complexes [29].
As is shown in Fig. 1, NO3 ligand coordinated as a monodentate ligand in aluminum and gallium complexes but as a bidentate ligand in indium complex. Therefore, the two first complexes were a five coordinated and the third one was six coordinated. Also, bond length of nitrate oxygen in indium complex was longer than gallium and aluminum. Optimized structure suggested a square planar geometry around central atom with two N atoms from diamine moiety and two O atoms of phenolic groups. Therefore, the axial position is occupied by nitrate group.
The frontier molecular orbitals played an important role in the reactivity of the compounds and in many electric and optic properties [30]. The difference of the energies of the HOMO and LUMO; the band gap; served as a measure of the excitability of the molecule, the smaller the energy easily it would be excited. The 3D plots of the HOMO and the LUMO of complexes under study are shown in Fig. 2. Large HOMO–LUMO gap automatically means high excitation energies for many excited states, a good stability and a high chemical hardness for the complex. In Al and Ga complexes, LUMO is on aromatic ring through empty π* orbitals, but in In complex LUMO is located on the nitrate.
Spectral analysis of complex–BSA interaction
UV–Vis study
The UV–Vis absorption spectra of BSA in the presence of various concentrations of gallium(III) complex show in Fig. 3. The absorbance of BSA increased with increasing concentration of the complex, which was obvious evidence for formation of the complex between BSA and the compound.
The absorption data were analyzed according to Eq. (1).
In this equation, ε a, ε f and ε b correspond to extinction coefficient of A observed/[BSA], the extinction coefficient for the free BSA and extinction coefficient for the complex in fully bound form, respectively. In the plot of [complex]/(|ε a − ε f|) versus [complex], K b is the ratio of the slope to the intercept (Fig. 4). K b values are listed in Table 3.
Fluorescence study
To interpret the data from fluorescence quenching studies, it is important to understand what kind of interaction takes place between the fluorophore (serum albumin) and the quencher (complex). Fluorescence quenching could proceed via different mechanisms usually classified as dynamic quenching, static quenching or both static and dynamic quenching participates in it simultaneously. Dynamic quenching mechanism depends upon diffusion, since higher temperatures result in larger diffusion coefficients, the quenching rate constants are expected to increase with increasing temperature. On the other hand, increasing temperature is likely to result in lower values of the static quenching constants [31]. A valuable feature of the intrinsic fluorescence of serum albumins is the high sensitivity of tryptophan and tyrosine to their local environment. Transformations in fluorescence spectra of tryptophan and tyrosine are common in response to protein conformational transitions, subunit association, substrate binding, or denaturation [32]. So, the intrinsic fluorescence of serum albumins can provide considerable information about their structure and dynamics, and is often considered in the study of protein folding and association reactions. The effect of Na2[Ga(5-SO3-1,2-salophen)NO3] on BSA fluorescence intensity is shown in Fig. 5. It was obvious that BSA had a strong fluorescence emission peak around 342 nm after being excited at 280 nm. When a fixed concentration of BSA was titrated with different amounts of Na2[Ga(5-SO3-1,2-salophen)NO3] in 0.001 M Tris–HCl buffer (pH = 7.4), a remarkable intrinsic fluorescence decreasing of BSA was observed. Similar situations were observed for the other complexes.
The fluorescence quenching data are analyzed by the Stern–Volmer Eq. (2):
where F 0 and F are the fluorescence intensities in the absence and in the presence of quencher, K SV is the Stern–Volmer quenching constant, and [Q] is the concentration of quencher (complex). The linear Stern–Volmer plots indicated that Eq. (2) is applicable for the present systems. The values of K SV represented the relative affinity of [M(L)NO3] for BSA. These results indicated that the probable quenching mechanism is dynamic quenching, because the K SV increased with increasing the temperature (Fig. 6). The plots of the left hand side of Eq. (3) versus [complex]/[BSA] (Fig. 7) confirmed the 1:1 stoichiometry between BSA and quencher.
The average BSA aggregation number, <J>, potentially induced by the complexes can be determined by using Eq. (3) (Table 4):
The slope, (<J>) <1, indicated that the complex binding did not induce any aggregation on BSA molecules and accordingly proved the 1:1 stoichiometry for complex:BSA systems (Fig. 7).
Determination of the binding constant and the binding site
The relationship between fluorescence quenching intensity and the concentration of quencher can be described by Eq. (4) [33]:
where F 0 and F are the fluorescence intensities in the absence and the presence of the complex, K b is the apparent binding constant of the complex with BSA, and n is the number of binding sites per BSA and [Q] is the quencher concentration (complex) (Fig. 8). These results showed that K b increased slightly with increasing the temperatures, but n is almost constant, which may be indicated that there was molecular binding between [M(L)NO3] and BSA according to 1:1 stoichiometry (Table 5).
Binding mode and determination of the thermodynamic parameters
The combining properties of the drug and proteins could be affected by various weak non-covalent interactions such as hydrophobicity, hydrogen bonding and stereo-effect. The sign and magnitude of the thermodynamic parameters such as ΔG (free energy change), ΔH (enthalpy change), and ΔS (entropy change) of the reaction were important for the study of the interaction forces. To obtain further insights into the weak interactions associated with the complexation of [M(L)NO3] with BSA, we tried to determine the thermodynamic parameters using the van’t Hoff equation (5) [34]:
where K b is the binding constant at the corresponding temperature and R is the gas constant. The van’t Hoff plots for this interaction are shown in Fig. 9. The enthalpy change (ΔH) is calculated from the slope of the van’t Hoff relationship and (ΔS) from the intercept. The free energy change (∆G) is calculated from Eq. (6) at T = 298 K:
All the thermodynamic parameters are listed in Table 5. Sign and magnitude of thermodynamic parameter showed host–guest systems that are associated with ∆H > 0 and ∆S > 0 are driven by the hydrophobic interactions, while those with ∆H ≈ 0 and ∆S > 0 by the electrostatic interactions and those with ∆H < 0 and ∆S < 0 by the hydrogen bonding or van der Waals interactions [35]. The negative sign of ∆G proved the spontaneity of the binding of [M(L)NO3] with BSA, the positive values of ∆H and ∆S show that the binding processes are mainly entropy driven. Thus, the interaction of the BSA with Ga complex is of hydrophobic mode. The same was valid for the other complexes.
Site-selective binding of the complexes on BSA by competition reaction between site markers and complexes
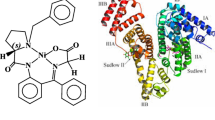
BSA has two main sites for binding to drugs: (i) site I of BSA which tryptophan residue is binding to drugs and (ii) site II which tyrosine is located (Fig. 10).
The former shows affinity for warfarin, phenylbutazone, etc., whereas the binding pocket of the latter is well suited for ibuprofen, diazepam, flufenamic acid, etc. To show the [ML] binding to tryptophan residue, we used phenylbutazone and ibuprofen as a well-known site marker for site I and site II, respectively. For this purpose, emission spectra of ternary mixtures of complex, BSA and site marker were measured. The results showed that the quenching of BSA in the presence of ibuprofen is very smaller than in its absence [36]. This has been typically shown in Fig. 11. Based on these results, we deduced that a competition existed for the binding of ibuprofen and complexes to site II. Therefore, the complexes binding to site II is inhibited by the simultaneous binding of the site II of ligand via ibuprofen, while it remained almost with no change by the addition of phenylbutazone. It could be concluded that the complexes were mainly located in the region of site II (subdomain IIIA) in BSA. These results were confirmed by molecular docking studies. The K b values in Table 6 are compared with the values in Table 5 for the absence and presence of site markers.
Energy transfer between the complexes and BSA
In Fig. 5, it also could be found that the fluorescence intensity of BSA was increased gradually at 430 nm and decreased at 340 nm, with an isosbestic point at 380 nm. These results indicated that a non-radiative energy transfer process took place through dipole–dipole coupling effect between BSA and the complex.
There was a spectral overlap between the fluorescence emission spectrum of the free BSA and absorption UV–Vis spectra of the complexes (Fig. 12). As the fluorescence emission of protein was affected by the excitation light around 280 nm, the spectrum ranging from 300 to 500 nm was chosen to calculate the overlapping integral.
The energy transfer efficiency between [M(L)NO3] and BSA was studied according to the Förster’s energy transfer theory. The Förster theory shows that energy transfer is affected not only by the distance between the acceptor and the donor, but also by the critical distance of energy transfer (R 0), which can be calculated using Eq. (7) [37]:
where F and F 0 are the fluorescence intensities of BSA in the presence and absence of quencher, r the donor–acceptor distance and R 0 is the critical distance where the transfer efficiency is 50 %:
In Eq. (8), K 2 is the space factor of orientation, N the refractive index of the medium, Φ the fluorescence quantum yield of the donor, J is the effect of the spectral overlap between the emission spectrum of the donor (BSA) and the absorption spectrum of the acceptor (complex), which could be calculated in Eq. (9):
where F(λ) is the fluorescence intensity of the donor and ε(λ) is the molar extinction coefficient of the acceptor. Figure 12 shows the overlap of the UV–Vis absorption spectrum of [Ga(L)NO3] with the fluorescence emission spectrum of BSA, K 2 = 2/3, N = 1.36 and Φ = 0.15 for BSA. Since the average distance of donor to acceptor is 2 < r < 8 nm and 0.5R 0 < r < 1.5R 0, the energy transfer from BSA to [Ga(L)NO3] occurred with high probability (Table 7).
Effect of the complexes on the protein conformation
It was observed that the fluorescence of serum albumins comes from the tyrosine, tryptophan and phenylalanine residues. The spectrum of BSA was sensitive to the microenvironment of these chromophores. To study molecular environment surrounding of BSA, chromophore synchronous fluorescence spectra could be used. The changes of maximum emission wavelength of the tryptophan residues will reflect the conformation changes of BSA [38].
Distinction of the difference between excitation wavelength and emission wavelength (∆λ) reflects the spectra of disparate chromophores. Large ∆λ values such as 60 nm indicated that the synchronous fluorescence of BSA was characteristic of tryptophan residue and those with small ∆λ values such as 15 nm was characteristic of tyrosine. The tyrosine residues and the tryptophan residues of the fluorescence spectra of BSA at various concentrations of [Ga(L)NO3] are shown in Fig. 13a and b, respectively. It is apparent from Fig. 13 that the emission peak of the tryptophan residue (Δλ = 60 nm) showed a gradual quenching and a faint red shift, indicating that the complex was located in close proximity to the Trp residues and the polarity near the tryptophan residues was increased and so the hydrophobicity was decreased faintly. When the hydrophobic part of the complex was close enough to the phenyl moiety of Trp, hydrophobicity near the tryptophan residues decreased involuntarily. But the emission peak of the tyrosine residue (Δλ = 15 nm) showed quenching without significant shift, which indicated that mentioned complex was close to Tyr residue but had no effect on the conformation of tyrosine microenvironment. Also, the quenching of the fluorescence intensity of tyrosine residues was stronger than that of the Trp residue, suggesting that Tyr residues contributed greatly to the quenching of the intrinsic fluorescence. Similar spectral feature were observed for the interaction of the other complexes with BSA.
Molecular docking study
The BSA molecule is made up of three homologous domains (I, II, III) that are divided into nine loops (L1–L9) by 17 disulfide bonds. Each domain is composed of two sub-domains (A and B) (Fig. 10). Intrinsic fluorescence of BSA is due to the presence of two tryptophan residues (Trp-134 and Trp-212). Trp-134 is located on the surface of subdomain IB and Trp-212 located within the hydrophobic binding pocket of subdomain IIA [39]. In this study, the MVD programs [25] were chosen to examine the binding mode of the complexes at the active site of BSA. The docking results showed that the complexes prefers the binding pocket of site II (subdomain IIIA), demonstrating that in free energy point of view the adduct formed by binding of the complexes to subdomain IIIA is more stable than that formed via interaction of the complexes to subdomain IIA. It should be noted that the grid box of docking was taken large enough so that it span both binding pockets of IIA and IIIA. However, the binding of the complexes to site IIIA produced lower free energy of binding. As it is obvious from Fig. 14, the complexes are situated within the subdomain IIIA cavity. Furthermore, there are also a number of specific hydrogen bonds, due to the presence of several polar residues in the proximity of the protein, which can play an important role in binding of the complexes to BSA via H-bonds. In fact, hydrogen bonding interactions can occur between the O, N, H of the Schiff base complex and the aminoacid residues of BSA (Fig. 14). The results suggested that the formation of hydrogen bonds stabilizes the complex–BSA. The binding energy for the binding of complexes to BSA found to be −126.79, −130.03, −131.00 kJ/mol for [Al(L)NO3], [Ga(L)NO3] and [In(L)NO3], respectively. These results are very close to those obtained by the above-mentioned experimental method.
Aminoacid residues involved in complexes–BSA interactions and binding energy for the best selected docking positions are listed in Table 8. It is important to note that the interactions between Schiff base complexes and BSA were not exclusively hydrophobic in nature since some of the residues are in the vicinity of the bound complex.
Conclusions
Three new water-soluble aluminum(III), gallium(III) and indium(III) complexes were synthesized and the interaction with BSA was investigated by various methods. A fluorescence method for the rapid and simple determination of the interaction between the complexes and BSA, as an important carrier protein, under simulated physiological condition of pH 7.4 was provided. The experimental results indicated that the intrinsic fluorescence of BSA can be quenched by complexes through the dynamic quenching mechanism; the binding reaction is spontaneous. The binding constants K b, at 298, 304 and 310 K, were determined according to a Stern–Volmer equation. Also, it was concluded that complex binding did not induce any aggregation on BSA molecules and accordingly proved the 1:1 stoichiometry for [M(L)NO3]:BSA systems. Experimental results by synchronous fluorescence spectra also showed that the microenvironment around the tyrosine residues did not show obvious changes during the binding process. Thermodynamic results indicated that the binding process was endothermic and essentially entropy driven suggesting that hydrophobic interactions play a significant role in the adduct formation. The high affinity of BSA for these complexes is clearly evidenced by ∆G values which clarified the role of the albumin as endogenous carrier for these complexes in the body. The distance (r) between BSA and complex, based on the Förster energy transfer theory, was calculated and indicated that energy transfer was highly possible. The independent docking studies resulted in binding constant and binding mechanism very close to the experimental ones. As the aim of this study was the effect of different metals on the bonding propensity of the complexes to BSA, K b values compared in this respect. The order of the K b values was as follows: [In(L)NO3] > [Ga(L)NO3] > [Al(L)NO3]. So, it is concluded that In(III) complex had more propensity to interact with BSA than Ga(III) and Al(III). Thus, the results are of great importance towards further designing and developing metal(III) complexes and assessment of BSA binding as drugs.
References
C.M. da Silva, D.L. da Silva, L.V. Modolo, R.B. Alves, M.A. de Resende, C.V.B. Martins, A. Fatima, J. Adv. Res. 2, 1 (2011)
S.H. Etaiw, D.M. Abd El-Aziz, E.H. Abd El-Zaher, E.A. Ali, Spectrochim. Acta A 79, 1331 (2011)
C. Metcalfe, J.A. Thomas, Chem. Soc. Rev. 32, 215 (2003)
P. Caravan, Ch. Orvig, Inorg. Chem. 36, 236 (1997)
N.C. Bhardwaj, S.C.S. Jadon, R.V. Singh, Synth. React. Inorg. Met. Org. Chem. 24, 1267 (1994)
H. Sugimoto, Ch. Kawamura, M. Kuroki, T. Aida, Sh. Inoue, Macromolecules 27, 2013 (1994)
S. Kashaniana, M.B. Gholiv, F. Ahmadia, A. Taravatia, A. Hosseinzadeh Colagar, Spectrochim. Acta A. 67, 472 (2007)
S. Gündüz Beniz, E. Maltas, S. Yıldız, Anal. Lett. 43, 2892 (2010)
V. Vajpaee, Y.P. Singh, J. Coord. Chem. 61, 1622 (2008)
Y. Sun, C.J. Anderson, T.S. Pajeau, D.E. Reichert, R.D. Hancock, R.J. Motekaitis, A.E. Martell, M.J. Welch, J. Med. Chem. 39, 458 (1996)
S.E. Harpstrite, A.A. Beatty, S.D. Collins, A. Oksman, D.E. Goldberg, V. Sharma, Inorg. Chem. 42, 2294 (2003)
V. Sharma, D.P. Worms, Chem. Rev. 99, 2545 (1999)
P.F. Salas, Ch. Herrmann, C. Orvig, Chem. Rev. 113, 3450 (2013)
T.J. Wadas, E.H. Wong, G.R. Weisman, C.J. Anderson, Chem. Rev. 110, 2858 (2010)
A. Sułkowska, J. Rόwnicka, B. Bojko, W. Sułkowska, J. Mol. Struct. 651, 133 (2003)
M. Asadi, Z. Asadi, S. Barzegar Sadi, R. Yousefi, A. Barzegar Sadi, H. Khalili, J. Coord. Chem. 65, 722 (2012)
J. Gao, Y. Guoa, J. Wang, Z. Wang, X. Jin, C. Cheng, Y. Li, K. Li, Spectrochim. Acta A. 78, 1278 (2011)
G. Fanali, Y. Cao, P. Ascenzi, M.J. Fasano, Inorg. Biochem. 117, 198 (2012)
B. Kumar Paul, A. Samanta, N. Guchhait, J. Phys. Chem. B. 114, 6183 (2010)
P. Panneerselvam, R.B. Nair, G. Vijayalakshmi, E.H. Subramanian, S.K. Sridhar, Eur. J. Med. Chem. 40, 225 (2005)
P. Kalck, F. Monteil, Adv. Organomet. Chem. 34, 219 (1992)
G. Zhang, A. Wang, T. Jiang, J. Guo, J. Mol. Struct. 891, 93 (2008)
E.A. Brustein, N.S. Vedenkina, M.N. Irkova, Photochem. Photobiol. 18, 263 (1973)
G.A. Petersson, M.A. Allaham, J. Chem. Phys. 94, 6081 (1991)
B. Hemmateenejad, M. Shamsipur, F. Samari, T. Khayamian, M. Ebrahimi, Z. Rezaei, J. Pharm. Biomed. 67–68, 201 (2012)
V.A.J. Aruna, V. Alexander, J. Chem. Soc., Dalton Trans. 9, 1867 (1996)
Y. Zhang, L. Ruan, W.J. Zhao, X.J. Wang, Z.A. Zhu, Polyhedron 22, 1535 (2003)
M.A. Mokhles, J. Chin. Chem. Soc. 48, 153 (2001)
C.R. Wade, B.J. Lamprecht, V.W. Day, J.A. Belot, Polyhedron 26, 3286 (2007)
E. Kose, A. Atac, M. Karabacak, P.B. Nagabalasubramanian, A.M. Asiri, S. Periandy, Spectrochim. Acta A 116, 622 (2013)
A. Sułkowska, J. Mol. Struct. 614, 227 (2002)
Y.J. Hu, W. Li, Y. Liu, J.X. Dong, S.S. Qu, J. Pharm. Biomed. Anal. 39, 740 (2005)
Y.J. Hu, Y. Liu, A.X. Hou, R.M. Zhao, X.S. Qu, S.S. Qu, Acta Chim. Sin. 62, 1519 (2004)
F. Samari, B. Hemmateenejad, M. Shamsipur, M. Rashidi, H.R. Samouei, J. Inorg. Chem. 51, 3454 (2012)
M. Asadi, Z. Asadi, L. Zarei, S. Barzgar Sadi, Z. Amirghofran, Spectrochim. Acta A 133, 697 (2014)
G. Zhang, L. Wang, P. Fu, M. Hu, Spectrochim. Acta A 82, 424 (2011)
Y.J. Hu, Y. Liu, Z.B. Pi, S.S. Qu, Bioorg. Med. Chem 13, 6609 (2005)
Y.J. Hu, Y. Liu, L.X. Zhang, J. Mol. Struct. 750, 174 (2005)
J. Ao, L. Gao, T. Yuan, G. Jiang, Chemosphere 119, 590 (2015)
Acknowledgments
We are grateful to Shiraz University Research Council for its financial support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Asadi, Z., Asadi, M. & Shorkaei, M.R. Synthesis, characterization and DFT study of new water-soluble aluminum(III), gallium(III) and indium(III) Schiff base complexes: effect of metal on the binding propensity with bovine serum albumin in water. J IRAN CHEM SOC 13, 429–442 (2016). https://doi.org/10.1007/s13738-015-0751-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13738-015-0751-4