Abstract
Autism spectrum disorder (ASD) research has yet to leverage “big data” on the same scale as other fields; however, advancements in easy, affordable data collection and analysis may soon make this a reality. Indeed, there has been a notable increase in research literature evaluating the effectiveness of machine learning for diagnosing ASD, exploring its genetic underpinnings, and designing effective interventions. This paper provides a comprehensive review of 45 papers utilizing supervised machine learning in ASD, including algorithms for classification and text analysis. The goal of the paper is to identify and describe supervised machine learning trends in ASD literature as well as inform and guide researchers interested in expanding the body of clinically, computationally, and statistically sound approaches for mining ASD data.
Similar content being viewed by others
Over the past two decades, there has been a significant rise in the generation, acquisition, and storage of data. The era of “big data” is marked by both a substantial increase in the rate at which data is generated and the variety of data produced. The growth in volume, velocity, and variety of data is due in large part to the availability and affordability of assorted instruments and infrastructure, used to collect and analyze many different types of information. Furthermore, the availability of various machine learning toolkits, such as Hadoop, TensorFlow, Spark, and R, combined with specialized hardware technologies has led to unique opportunities for researchers to leverage machine learning algorithms. This rise in computing power and processing technologies has opened the door for the application of machine learning theories in different fields of study.
One such field is autism spectrum disorder (ASD) research. ASD is a neurodevelopmental disorder characterized by deficits in social communication and social interaction, in addition to restricted, repetitive patterns of behavior (American Psychiatric Association 2013). The Centers for Disease Control and Prevention (CDC) currently estimates that one in 59 children is diagnosed with ASD in the USA (Autism and Developmental Disabilities Monitoring Network 2016). This may be an underestimate given the results of a recent parent survey finding a prevalence rate of one in 45 (Zablotsky et al. 2015). ASD is a heterogeneous disorder with diversity observed with respect to symptom presentation and severity, risk factors and etiology, as well as treatment response (Lord et al. 2000a). The high prevalence rate and heterogeneous nature of ASD have led some researchers to turn to machine learning over traditional statistical methods for data analysis.
Machine learning can be broadly sorted into two categories, unsupervised and supervised learning. This paper focuses on the latter of these approaches. Supervised machine learning involves algorithms that use input variables to predict a target classification (i.e., the dependent variable), which may be categorical or continuous. Unlike unsupervised learning (clustering), supervised learning involves datasets where the target prediction (e.g., diagnosis) is known at training time for the data used to learn the model. A supervised learning model is deemed successful when the model can (a) accurately predict the target result for a training dataset to a certain degree of accuracy and (b) be generalized to new datasets beyond those used to train the model. To improve a model’s ability to make predictions on future data, a method called cross-validation is often employed. This method allows a subset of the data to be removed before training, so that the model can then be tested on “new” data. A K-fold cross-validation strategy separates the available data into K-subsets and trains the model on all but one of the subsets and tests on the remainder. The process is repeated until the model has been trained on all the available data. The performance scores across the runs are averaged. In a leave-one-out cross-validation (LOOCV) method, all but one data point is used to train the model, which is then evaluated on the held out point. This process is repeated for each of the data points. A supervised machine learning model’s success is typically measured according to accuracy (i.e., the ability to correctly classify into separate categories). This may be further broken down to consider sensitivity (i.e., the ability to correctly detect true positives) and specificity (i.e., the ability to correctly detect true negatives). A further measurement of a supervised machine learning model’s success is AUC or area under the receiver operating character curve (ROC). The ROC is a plot of sensitivity vs specificity, and the area under the curve depicts how well a method makes positive and negative categorical distinctions.
This survey contributes to the understanding of ASD research as a whole, since it sheds light on new methods that allow researchers to take advantage of the larger datasets that are increasingly available to them. Exposing researchers to a survey of these new methods both helps increase their understanding of how and where the field is expanding and helps summarize the conclusions made using these methods on large datasets. In addition, by providing a comprehensive review of existing supervised machine learning research in ASD, we hope to assist other researchers in finding areas in this domain that can benefit from the techniques described here.
Method
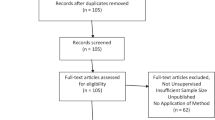
Endnote and Google Scholar were searched for peer-reviewed articles using combinations of the search terms “autism,” “ASD,” “pervasive developmental disorder,” “PDD,” “analytics,” “data science,” “diagnosis,” “genetics,” “supervised learning,” “machine learning,” “text mining,” “data mining,” and “literature mining.” Searches were limited to articles published between 2005 and 2018 and met the requirements of the PRISMA statement (Appendix A) (Moher et al. 2009).
The abstracts, methods, and results sections of the articles were reviewed by a team of five researchers with expertise in computational and data science (machine learning) as well as autism spectrum disorder, with each article receiving at least two independent reviews. Using a standardized rubric, an article was included if the following criteria were met: (a) was published in a peer-reviewed journal; (b) included a group of individuals with ASD, autistic disorder, Asperger’s syndrome, or pervasive developmental disorder-not otherwise specified (PDD-NOS); and (c) a supervised machine learning model was utilized as the primary method of analysis. While these procedures may not have produced an exhaustive review, findings should be representative of the current trends of machine learning in the field of ASD.
Results
The literature search produced 27 publications from Endnote and 94 from Google Scholar. After abstracts and methods were reviewed, 45 research articles were included. Of the included articles, 35 involved the use of supervised machine learning in original ASD research and 10 involved the use of supervised machine learning in text mining of existing ASD literature.
Supervised Machine Learning in ASD Research
The supervised learning research reviewed in this paper includes classification algorithms designed to identify patterns in a given dataset that will lead to a correct diagnosis or other classification of participants. Many of the studies described in this review applied a number of different supervised machine learning methods, with one or more superior performing models emerging from the group. Of the 35 articles included in this review, with a number of studies reporting success with various models, support vector machine (SVM) algorithms were successfully used in 13 studies, applied alternating decision tree (ADTree) methods were used in six, least absolute shrinkage and selection operator (LASSO) regression was used in four, random forest (RF) was used in four, neural networks were used in three, ridge regression was used in four, deep learning was used in two, elastic net regression (ENet) was used in two, linear discriminant analysis was used in two, logistic regression was used in two, decision tree were used in two, conditional inference forest (CF) was used in one, decision stump was used in one, flex tree was used in one, naïve Bayes was used in one, and random tree was used in one. Each of these methods will be discussed in turn. Refer to Table 1 for a summary of articles and the machine learning methods used.
Support Vector Machines
An SVM is a supervised learning algorithm that fits an optimal hyperplane in an n-dimensional space to correctly categorize the target result using the independent variables in the dataset. An SVM is a maximum margin classifier, meaning it maximizes the separation between n classes of data effectively in a high-dimensional space (Bishop 2006). SVMs are especially useful when the boundary between groups is non-linear because points can be easily transformed to a space in which the boundary is linear (Bishop 2006). Because of this feature, SVMs are generally used in classification problems in which the distinction between groups is non-linear. SVM algorithms have been used in the research included in this review to classify individuals (e.g., according to diagnosis) based on standardized assessments, genes, neuroimaging, and other measurements.
Behavioral and Neuroimaging Data
Machine learning algorithms—such as SVMs—have been used in ASD research in attempts to improve screening and diagnostic practices. In general, these models have been shown to improve the accuracy of diagnoses and provide insight into how different characteristics (such as standardized assessments, eye movement data, upper limb and general kinesthetic data, and neuroimaging data) can help distinguish between patients with and without ASD. Currently, standardized assessments reliant on behavioral observation are the clinical standard for diagnosing ASD. One limitation of standardized assessments is the potential for misdiagnoses, particularly when distinguishing one disorder from another. Bone et al. (2016) trained and cross-validated an SVM classifier to differentiate ASD from other developmental disorders (DD) based on data from two standardized assessments, the Autism Diagnostic Interview, Revised (ADI-R; Le Couteur et al. 2003) and the Social Responsiveness Scale (SRS; Constantino and Gruber 2007). The sample consisted of 1264 individuals with ASD and 462 individuals with a DD other than ASD. Participants were segmented into two groups: individuals 10 years of age or older and individuals under 10 years of age. Using only five behavioral codes, results of the SVM screen algorithm showed a sensitivity of 89.2% for individuals 10 years of age or older and 86.7% for individuals under 10 years old. A specificity of 59.0% was reported for individuals 10 years of age or older and 53.4% for individuals under 10 years old. Given the potential for standardized assessments to fail to distinguish one disorder from another, these findings suggest that machine learning may be used to improve accuracy of standardized assessments.
Machine learning models have also been employed to evaluate potential screening tools, alternative to behavioral assessments, including computer-aided methods. Liu et al. (2016) used an SVM classification algorithm to explore whether patterns of eye movement during face recognition tasks can differentiate children with ASD from children with typical development (TD). Participants included 29 children with ASD and 29 children with TD ages 4 to 11 years old. Using data recorded from eye tracking software, an SVM classifier model was generated using a LOOCV strategy to train the model and test its ability to detect ASD. Feature representation was conducted in two parts. The SVM classifier was trained based on the extracted histograms containing the visual attention information on each face area (defined by K clusters). The results of the analysis revealed an accuracy of 88.51% in discriminating participants with ASD from participants with typical development, with a sensitivity of 93.10%, a specificity of 86.21%, and an AUC of 0.8963. Furthermore, the top seven of 64 discrimination dimensions were found to produce a relatively strong accuracy of 79.31%, with a sensitivity of 68.96% and a specificity of 84.48%. These findings reveal preliminary support for the application of machine learning analysis of face scanning patterns as a computer-aided screening method for the detection of ASD.
Similarly, Crippa et al. (2015) evaluated a potential computer-aided screening method for ASD using an SVM classifier. The authors set out to investigate whether upper limb movement could accurately differentiate low functioning children with ASD from children with TD. A total of 15 preschool-aged children with ASD and 15 children with TD, matched on developmental age, participated in the study. An SVM was used to classify participants. Using seven movement-related features, the method produced a maximum classification accuracy of 96.7%, with a sensitivity of 100% and a specificity of 93.8%. Leveraging LOOCV, the overall mean classification revealed an accuracy of 84.9%, with a specificity of 89.1% a sensitivity of 82.2%. Findings suggest that machine learning analysis of kinematic data may detect motor features that are useful in identifying at least a portion of individuals with ASD (i.e., low functioning and preschool aged).
Kinematic data was also used by Li et al. (2017) in a model to distinguish adults with ASD from adults with TD. Participants included 16 adults with ASD and 14 adults with TD, matched on IQ. Based on the performance of hand movement tasks, a naïve Bayes classifier of the means and standard deviations of 20 kinematic parameters was used to choose which of the eight imitation conditions were most useful to discriminate between subjects with ASD and subjects with TD. Then, an SVM was used on the chosen parameters. The performance of the SVM was lower than the accuracy of the naïve Bayes classifier, which the authors speculate was due to overfitting. Four models were then tested, including an SVM, RF, naïve Bayes, and decision tree. Both the naïve Bayes and SVM models outperformed the other models on average. A linear SVM achieved the highest accuracy of 86.7%, with 85.7% sensitivity and 87.5% specificity.
Voice prosody was examined by Nakai et al. (2017), comparing the performance of machine learning vs the clinical judgment of speech therapists in classifying children with ASD and children with TD based on single-word utterances. Participants included 30 children with ASD and 51 children with typical development. All children were between the ages of 3 and 10 years old and had no comorbid disorders. After isolating their single-word responses, an SVM classifier with cross-validation on 24 features was employed to identify ASD or TD. The SVM proved more accurate (76%) than the 10 speech therapists whose classifications were also based on the same audio recordings (69%). The SVM had a sensitivity of 81% and specificity of 73% compared to the therapists, who demonstrated a sensitivity of 54% and specificity of 80%. This study shows the potential of machine learning analysis of speech prosody as a useful screening tool.
Neuroimaging data has also been examined using machine learning approaches. Moradi et al. (2017) predicted symptom severity of individuals with ASD based on cortical thickness using support vector regression (SVR) and ENet penalized linear regression. Participants included 156 individuals with ASD ages 8 to 40 years old. The dataset was compiled from four sites in the Autism Brain Imaging Data Exchange (ABIDE; Di Martino et al. 2014). ASD severity was based on the participants’ Autism Diagnostic Observation Schedule (ADOS; Lord et al. 2000b) scores. As compared to SVM, SVR uses the same mathematical representation of the input data, but the output is continuous instead of discretized. In SVR, a regression line is fit to the data with a tube of predetermined radius. A total of 78 outputs from the SVR were used as the input for an ENet linear regression to produce the desired symptom severity scores. The authors used 10-fold cross-validation and found an average correlation of 0.51 and an average mean absolute error of 1.36. These results show an improved correlation score across multiple sites over previous studies and show the potential of machine learning in identifying biomarkers linked to behavioral symptoms of ASD.
Zhang et al. (2018) sought to identify male children with ASD from TD children though diffusion magnetic resonance imaging (dMRI). Using the data of 70 children diagnosed with ASD and 79 typically developed controls obtained from the Center for Autism Research through Children’s Hospital of Philadelphia, this study analyzed their whole brain white matter connectivity with the help of a SVM and 10-fold cross-validation. By extracting multiple diffusion features from each fiber cluster of each subject, they were able to classify subjects as ASD or TD. The model with the highest accuracy, 78.33%, occurred with 4697 valid fiber clusters. This model produced a sensitivity of 84.81% and specificity of 72.86%. This study found that the most discriminatory fiber tracks (corpus callosum, cerebellum, brain stem, uncinate fasciculus, arcuate fasciculus, superior longitudinal fasciculus, and aslant) correspond to the fiber tracks identified with ASD in previous research. Using machine learning allowed Zhang and colleagues to better study locally specific brain regions via white matter parcellation into a large number of fiber clusters.
Behavioral and Developmental Data
Differentiated from many studies, Bussu et al. (2018) used longitudinal data captured at multiple points in development (8 and 14 months of age) for high-risk siblings to increase accuracy of predicting ASD diagnosis at 36 months. The behavioral measures used include Mullen Scales of Early Learning (MSEL; Mullen 1995), Vineland Behavioral Scales (VABS; Sparrow et al. 1984), and the Autism Observation Scale for Infants (AOSI; Bryson et al. 2008). A total of 161 high-risk siblings were separated into three groups: HR-ASD (n = 32), HR-Atypical (n = 43), and HR-Typical (n = 86). Two binary classification targets were created from the three groups where the final classification is either HR-ASD vs (HR-Atypical + HR-Typical) or (HR-ASD + HR-Atypical) vs HR-Typical. A lease squares SVM was used to train the classifiers with a 10-fold cross validation to tune model parameters. Based on the highest AUC, the classifier with the best performance for HR-ASD vs (HR-Atypical + HR Typical) at 14 months uses the Daily Living Score with an AUC of 71.3 which in comparison to data at 8 months having an AUC of 65.1 using motor scores, while the classifier using data at 8 months and the change factor leveraged motor, social, and daily living scores resulting an AUC of 65.9 which is a slight improvement over the 8-month scores. The best classified of (HR-ASD + HR-Atypical) vs HR Typical is also using data at 14 months but uses VABS scores and AOSI total score with an AUC of 70.8, in comparison the data at 8 months has an AUC of 69.2 using motor and communication scores, while the classifier using data at 8 months and the change factor had an AUC of 69.4. For this classifier, the performance did not differ from an AUC of 70 substantially as we saw in the HR-ASD vs (HR-Atypical + HR Typical) modeling. This longitudinal study allows for predictive measures to be used at their individual time points but also the observed change between 8 and 14 months.
Genetic Data
Machine learning has also been applied in ASD genetics research. SVMs allow researchers to determine which genes are related to ASD. These findings highlight the potential of machine learning to improve understanding of the role genes play in the development of ASD and other disorders. The identification of genes associated with ASD helps to shed light on the etiology of the disorder. Using curated lists of genes known to be associated with ASD and intellectual disability (ID), Kou et al. (2012) employed supervised machine learning techniques to classify known and predict new genes linked to these diagnoses. Computational methods, including two network-based classifiers and one attribute-based classifier, were employed. Finally, 10 SVM classifiers were employed using positive gene sets (i.e., genes associated with ASD or ID) and negative sets, which were randomly generated with 200 genes in each set. The SVM performed better than both network-based classifiers. Two non-overlapping lists were generated for truth data, containing 114 known rare, high-risk genes associated with ASD and 223 genes associated with ID. The SVM classifiers were capable of discriminating between genes associated with ASD, genes associated with ID, and other genes with an accuracy of 80 to 98%. Sensitivity for the classifiers ranged from 76 to 89%, specificity ranged from 89 to 96%, and AUC ranged from 0.94 to 0.97. Furthermore, the ASD classifiers performed better than ID classifiers.
Krishnan et al. (2016) also used an SVM to find potential genes correlated with ASD. A brain-specific functional network was created using a regularized Bayesian model. The network was used to identify 594 genes that may be predictors for ASD with varying levels of evidence (E1-E4). SVM models were trained and tested using 5-fold cross-validation in order to classify a patient with ASD or TD. Their best combination of phenotypes performed fairly, attaining an AUC of 0.80 using only E1 (high confidence genes). All evidence-weighted classifiers outperformed their unweighted counterparts.
Alternating Decision Tree
ADTree models, also known as acyclic directed graphical models, involve combining many one-level decision trees (i.e., decision stumps) to obtain a representation in which each stump consists of a decision node and two prediction nodes (Williams 2011). One main benefit of leveraging a decision tree is being able to interpret and understand the impact of each variable in the tree. While other methods, such as an RF, may lead to better performance from a predictability standpoint, they may lack the transparency that a decision tree gives. Decision trees can also be leveraged to identify unique segments in a population where multiple models can be applied rather than only a single model for the entire population. ADTree models have been used in the reviewed ASD research in attempts to enhance diagnostic and screening practices.
Behavioral and Neuroimaging Data
As mentioned previously, ASD is currently diagnosed via standardized behavioral assessments, which can be lengthy and time consuming to administer. In an attempt to accelerate the diagnostic process, Wall et al. (2012a) set out to identify a subset of ADI-R items that could be used to accurately classify ASD. A dataset of 891 individuals with ASD and 75 individuals without ASD who completed the ADI-R was initially tested. Using seven of the 93 ADI-R items, an ADTree classifier was found to perform best with an accuracy of 99.9%. The seven-item classifier was further examined in two additional samples of individuals with ASD (n = 1654; n = 322). Across both samples, the classifier was found to have an accuracy of nearly 100%. Specificity was evaluated using two small samples of individuals without ASD (n = 5; n = 12) as well as 1000 artificially generated score sheets used as control data, with specificity ranging from 93.8 to 99%, for both real and artificial data.
In a similar study, Wall et al. (2012b) set out to identify a subset of items included in the ADOS that could be used to accurately detect ASD. An initial sample including 612 individuals with ASD and 15 individuals without ASD who had completed module one of the ADOS were tested. Using eight of the 29 items measured in module one of the ADOS, an ADTree classifier again demonstrated a superior performance with a classification accuracy of 100%. The eight-item classifier was further validated using two additional samples of individuals with ASD (n = 110; n = 366) and 1000 artificial controls. Across both ASD samples, the eight-item classifier was found to have a sensitivity of 100%. Using the simulated control data, a specificity of 94% was revealed. The findings of these two studies by Wall and colleagues show promise in the use of machine learning techniques to sizably reduce the number of behavioral assessment items required to accurately classify ASD.
In a replication study, Bone et al. (2015) discussed the methodological issues that they felt raised concern in the studies conducted by Wall and colleagues (Wall et al. 2012a; Wall et al. 2012b). The primary concern was the unbalanced datasets used to test and validate the classifiers, which contained little to no negative cases (i.e., participants without ASD). The authors conducted experiments to replicate the results of Wall and colleagues’ ADTree procedures using larger, though not publicly available, datasets with more balanced samples of positive and negative cases. Both experiments failed to replicate the level of accuracy achieved by Wall and colleagues for both the reduced-item ADI-R and ADOS classifiers. Furthermore, the authors did not find that clinician administration time decreased using the reduced-item classifiers.
These classifiers have undergone additional evaluation for use as preclinical screening tools to detect risk of ASD. Duda et al. (2014) set out to further validate the eight-item ADOS classifier, referred to as the observation-based classifier (OBC). The study dataset included a more balanced sample of 2333 children with ASD and 283 children without ASD, who had completed the ADOS. The OBC, which uses an ADTree procedure to classify ASD, demonstrated significant correlations with both original ADOS and ADOS-2 (Gotham et al. 2007) scoring procedures, revealing a highest reported accuracy of 95.8%, sensitivity of 97.7%, and specificity of 83.5%. OBC outcomes were also compared to best estimate clinical diagnoses, showing an accuracy of 96.8%, sensitivity of 97.1%, and a specificity of 83.3%. While the results are not as robust as those initially reported by Wall et al. (2012b), the OBC may be a viable screening tool to detect risk of ASD.
The seven-item ADI-R classifier has also undergone further evaluation by Duda et al. (2016a). In this study, a mobile application, the Mobile Autism Risk Assessment (MARA), was used to administer the ADI-R questionnaire. Participants included 222 children and adolescents (69 of which were diagnosed with ASD), between the ages of 16 months and 17 years old, who were visiting a developmental-behavioral pediatric clinic for the first time. Participant’s MARA screening outcomes were compared to clinical diagnoses made during the visit. MARA utilizes an ADTree trained on the answer sheets of 891 subjects with ASD and 75 subjects without ASD from the Autism Genetic Resource Exchange (AGRE; Geschwind et al. 2001) database to generate scores. The model demonstrated a sensitivity of 89.9% and a specificity of 79.7%.
Supervised Feed-Forward Artificial Neural Networks
Neural networks are layered connections of neural units, or perceptrons, which can learn mappings between input data and desired outputs. The perceptrons typically make use of sigmoidal activation functions to accommodate non-linearity. Once a network is trained, new data can be input, and based on the learned weights and biases of each neural unit, an output will be generated. Neural networks are generally faster than other methods used for non-linearly separable problems (such as SVMs) (Bishop 2006). In the reviewed research, neural networks have been applied to behavioral assessments to detect ASD, to evaluate the impact of treatment intensity on treatment gains, and to study gene expression in ASD. In general, the application of Feed Forward Neural Networks (FFNNs) to ASD research has shown promising improvements both in diagnosis and in determining ASD phenotypes, often improving on existing methods.
Behavioral and Neuroimaging Data
Florio et al. (2009) set out to evaluate the accuracy of a neural network model in detecting ASD. The dataset included results of the Developmental Behavior Checklist (DBC; Einfeld and Tonge 1995), a parent-rated assessment that measures emotional and behavioral problems in children and adolescents with DD. The authors used a feed-forward neural network to predict whether or not a participant had ASD. The model was first tested in a sample of 319 individuals with ASD and 319 individuals a DD other than ASD. The model was then cross-validated using a separate sample of 62 individuals with ASD and 38 individuals with DD. In the initial test, the model revealed an accuracy of 92% and an AUC of 0.93. During cross-validation, the model produced an accuracy of 80%, sensitivity of 92%, specificity 70%, and AUC of 0.88.
Neural networks have also been leveraged by Linstead et al. (2015, 2017) in a regression task to learn the relationship between treatment intensity and learning outcomes in the context of applied behavior analysis (ABA) treatment for ASD. The authors found that, compared to simple linear regression, neural networks were more accurately able to model the relationship between treatment intensity and learning outcomes, explaining substantially more variance for a sample size of 726 patients. This work leveraged only shallow neural networks with a single hidden layer and considered patients only within the early intervention age range. Despite these limitations, this work highlighted the advantage of neural networks to learn non-linear relationships without any prior knowledge of the functional form of those relationships.
Genetic Data
Neural networks have also been used to explore gene expression. Xiong et al. (2015) used an ensemble of neural networks to analyze DNA sequences and predict to what extent genetic variants may disrupt RNA splicing, an essential element of gene expression that may contribute to various disorders including ASD. The genomes of five individuals with ASD and 12 controls were scanned for single nucleotide variations (SNVs) resulting in approximately 42,000 SNVs per subject. Each neural network in the ensemble had 2 layers and used 41,820 potential input-to-hidden parameters and 960 hidden-to-output parameters. To overcome the numerical difficulties of fitting a single model, a Bayesian Markov chain Monte Carlo technique was deployed. Markov chain Monte Carlo (MCMC) sampling methods allow sampling of multiple distributions, which will scale to the dimensionality of the sample space (Bishop 2006). The final model was used to score genetic variations. Of the 171 genes found to be misregulated in individuals with ASD, 27% were found to have high expression in the brain compared to only 13% of the 249 genes identified in the controls. The authors highlighted 19 genes potentially linked to ASD with known neurological, neurobehavioral, or neurodevelopmental phenotypes. While some of these genes have been previously associated with ASD, others were new candidate ASD genes.
Random Forest
An RF is an ensemble learning method that utilizes many individual classification and regression trees. A classification decision is made based on a majority vote of the trees predictions. A beneficial feature of using an RF is the built-in bootstrapping, which leads to training and validation of the algorithm with less intrinsic bias in the analysis (Mayor 2015). An RF is beneficial in uncovering non-linearities across a dataset that a simpler, more interpretable, generalized linear model may not uncover. One negative to using this ensemble method is the lack of easy interpretation due to the black box nature of the algorithm. Having the flexibility to vary the number of variables at each tree split and the number of trees to build allow the algorithm to find its optimal tuning parameters. The built in training and validation inherent in the algorithm will also have a positive effect on reducing the likelihood of overfitting to the training dataset. RF has been used in the reviewed ASD research to analyze neuroimaging data, monitor populations diagnosed with ASD, and develop screening tools. These methods have led to high diagnostic accuracy based on neuroimaging data, video data, and electronic health records. Again, these models often outperformed existing tools.
Behavioral and Neuroimaging Data
Chen et al. (2015) used machine learning models, including RF, to analyze neuroimaging data for diagnostic classification purposes. Low-motion resting-state functional MRI (rs-fMRI) scans were used for a sample of 126 individuals with ASD and 126 individuals with TD. Participants were matched based on age, non-verbal IQ, and head motion. Diagnostic classification was based on a matrix of functional connectedness between 220 identified regions of interest. Using the top 100 regions of interest, an RF produced the greatest level of accuracy of the models tested for diagnostic classification at 91%, with a sensitivity of 89% and a specificity of 93%. When applied to the top 10 regions of interest, the RF achieved an accuracy of 75%, with a sensitivity of 75% and a specificity of 75%. The high number of regions required to produce a strong accuracy in detecting ASD may imply that brain biomarkers for ASD are scattered rather than localized.
RF classifiers have also been used to develop affordable and efficient ASD screening tools. Abbas et al. (2018) combined two independent classifiers to create an ASD screening tool based on a parental questionnaire and behaviors observed in home videos. ADI-R and ADOS scoresheets from children ages 18 to 84 months were cultivated from multiple repositories and used to train an RF for a parental questionnaire and an RF video classifier, respectively, with clinical diagnosis as the target variable. To help balance the data, ADI-R interviews were administered to a random sample of low-risk children. Since language development often affects the presentation of ASD, the classifiers were trained and tested on children older and younger than 4 years old separately. The parental classifier was trained on 2299 children with ASD, 100 children with TD, and 287 children with another condition. The video classifier was trained on 3310 children with ASD, 585 children with TD, and 364 children with another condition. Once trained, the video classifier used the presence and severity of behaviors, as determined by analysts watching two to three 1-min semi-structured home videos, as inputs. Final diagnosis utilized logistic regression on the results of the questionnaire and video classifiers and performed better than other established screening tools, including the Modified Checklist for Autism in Toddlers (MCHAT; Robins et al. 2014) and the Child Behavior Checklist (CBCL; Achenbach and Rescorla 2001), based on the ROC curves.
Electronic Health Records
Bishop-Fitzpatrick et al. (2018) leveraged ICD-9 codes, V-codes, and E-codes from the electronic health records of 91 decedents with an ASD (or related) diagnosis and 6186 control decedents to build a random forest classifier. The goal of this study was not only to distinguish ASD from the control decedent but also to examine the lifetime health problems of those with ASD. From the first RF model, the top 50 ICD-9 codes, V-codes, and E-codes were chosen and were used to build a second, smaller random forest model. The model had an accuracy of 93%, sensitivity of 75%, specificity of 94%, and an AUC of 0.88. The 50 codes were then ranked in order of importance. The authors report that, overall, decedents with an ASD diagnosis have higher rates of nearly all 50 ICD-9 codes, V-codes, and E-codes.
Logistic Regression
Logistic regression is a generalized linear model used for binary classification, taking as input a set of independent variables. The independent variables can be discrete or continuous and are used to predict the probability or odds of the target result taking a particular binary value. To avoid overfitting the regression model, regularization techniques such as LASSO, ridge, and ENet can be deployed. LASSO, also known as L1 regularization, attempts to minimize the sum of the absolute value of the regression coefficients, which leads to automatic feature detection, removing input parameters that may provide little added value to the model. Ridge regression, or L2 regularization, constrains the sum of the squared differences of the regression coefficients, which minimizes an irrelevant feature’s effect on the regression model, but unlike LASSO will not actually remove it (Bishop 2006). The ENet regression combines the LASSO and ridge penalties (Zou and Hastie 2005). Logistic regression model literature shows attempts to improve ASD screening practices, analyze neuroimaging data, and evaluate risk factors for developing ASD. Linear discriminant analysis is similar to logistic regression, as it uses a linear combination of continuous, independent variables to predict a categorical outcome; however, linear discriminant analysis assumes that the independent variables are normally distributed.
ASD Risk Factors
Logistic regression has been used by Grether et al. (2009) to evaluate the impact of parental age on the risk of developing ASD. The study sample included 7,550,026 individuals born in California between 1989 and 2002. Data from the California Department of Developmental Services were used to identify individuals with ASD (n = 23,311) while the remainder of the sample made up the control group. Using maternal and paternal ages obtained from birth certificates, the authors applied logistic regression to determine risk of developing ASD, with race/ethnicity, parental education level, and year of birth taken into account as covariates. Increased maternal age by 10 years was found to elevate the risk of ASD by 38% compared to 22% for increased paternal age by 10 years. These findings persisted through all subgroups based on covariates, although it was found that these risks greatly increased among first-born children. These findings demonstrate the potential of machine learning to evaluate risk factors for developing ASD in very large populations.
Behavioral and Neuroimaging Data
Extending the work of Wall et al. (2012b), Kosmicki et al. (2015) set out to determine if a subset of the behaviors measured by the ADOS modules two and three could be used to accurately detect ASD. ADOS scores of 4540 individuals were included in the dataset. A total of 1451 individuals with ASD and 348 without ASD had completed module two of the ADOS and another 2434 individuals with ASD and 307 without ASD had completed module three. Unlike Wall et al. (2012b), ADTree was not found to be the best machine learning model tested. Using nine of the 28 behaviors measured within module two, a ridge regression performed best in correctly classifying participants with an accuracy of 98.27%, sensitivity of 98.81%, and specificity of 89.39%. An SVM (described above) was found to be the optimal method for module three. Based on 12 of the 28 behaviors measured, the SVM was able to classify participants with an accuracy of 97.66%, sensitivity of 97.71%, and specificity of 97.20%.
In a similar vein, Duda et al. (2016b) used machine learning models in another effort to produce a reduced-item classifier, this time using the SRS. Six machine learning models were trained and tested using a sample of 2775 individuals with ASD and 150 individuals with attention deficit hyperactivity disorder (ADHD) who had completed the SRS. Forward feature selection, under sampling, and 10-fold cross-validation were employed. Of the machine learning models tested, strong and comparable results were demonstrated by four models, including SVM, ridge regression, LASSO, and linear discriminant analysis. Each of these models relied on five of the 65 items measured by the SRS and revealed an AUC ranging from 0.962 to 0.965. These findings reveal that machine learning procedures can discriminate individuals with ASD from individuals with ADHD with a high level of accuracy.
Continuing their goal of developing an easy and widely available tool to distinguish between ASD and ADHD, Duda et al. (2017) collected and analyzed data from a web-based 15-question parent survey. Participants included 248 individuals with ASD and 174 individuals with ADHD, ages 2 to 17 years old, with no comorbidities base on parental report. A second archival dataset with SRS scoresheets was obtained from multiple repositories. The archival dataset included 2775 subjects with ASD and 150 subjects with ADHD. Subjects were diagnosed by a physician and had no comorbidities. The dataset was subsampled to maintain diagnosis proportions, and only the 15 features correlated to the survey were retained. Five machine learning models were selected based on the results of Duda et al. (2016b): SVM, LASSO regression, ridge regression, linear discriminant analysis, and ENet regression. Three independent experiments were performed with the models, including (1) training on archival data and testing on survey data, (2) training on survey data and testing on archival data, and (3) training and testing on mixed archival and survey data. The performance of each model varied greatly based on the training and testing sets, but all models performed best (AUC > .9) when the archival set was used from training. ENet and linear discriminant analysis performed significantly better than the other models when tested on survey data (AUC 0.89 ± 0.01). These models show potential for rapid risk score calculation available online.
Neuroimaging has also been examined using logistic regression. Plitt et al. (2015) explored the potential of rs-fMRI scans in identifying ASD biomarkers. Using a cohort of 59 individuals with high functioning ASD and 59 individuals with TD, the authors trained multiple models on three different regions of interest in fMRI scans along with demographic data, such as age, IQ, and SRS scores. A LOOCV was performed on each of the nine machine learning models explored across the three different regions of interest sets with ridge logistic regression (73.33% average accuracy) and linear SVM (73.89% average accuracy) outperforming the rest. When excluding the fMRI scans, the behavioral classifiers achieved as high as 95.19% accuracy. The authors also examined a dataset of 89 subjects with ASD and 89 subjects with TD from three ABIDE database sites. The ABIDE cohort performed markedly lower than the in house models. While machine learning analysis of rs-fMRI data shows promise in revealing ASD biomarkers, these results show it is not yet meeting the benchmark standards.
More recently, Rane et al. (2017) deployed a logistic regression model to build an open source whole brain classifier to predict ASD diagnosis. fMRI scans from the ABIDE database were used for 539 individuals with ASD and 573 individuals with TD. Features for a logistic regression model were selected from voxels within a gray matter template mask in the MNI152 space. LASSO regression and 5-fold cross-validation were utilized in an effort to avoid overfitting with the final model achieving 62% accuracy.
An additional use of a resting-state functional MRI (rs-fMRI) was done by Yamagata et al. (2018). Their work differentiated prior studies analyzing endophenotypes (measureable and heritable component between genes and disease diagnosis) that are present in both individuals with ASD and their non-ASD affected siblings by including an additional group of typically developing (TD) siblings. Enrolling additional pairs of TD siblings (with and without the ASD Endophenotype) controls for differences between TD siblings that would have previously been unaccounted for. The data consisted of a relatively small sample size of 60 male participants without intellectual disabilities, split in half between pairs of siblings with the ASD endophenotype (15 participants with ASD and 15 unaffected Siblings) and pairs of TD siblings (15 pairs of TD siblings without a family member having been diagnosed as having ASD). A sparse logistic regression was trained using Leave-one-pair-out cross validation (LOPOCV) (“pair” referring to pair of siblings) to reach a classification accuracy of 75% and an area under the curve of 78%. The study identified nine functional connections that in combination may serve as the potential endophenotype.
Deep Learning
Deep learning uses multiple hidden layers in a neural network architecture that mimic the brain’s neural connections. In the reviewed literature, deep learning has been used to study predictors of challenging behavior and analyze neuroimaging in individuals with ASD.
Behavioral and Neuroimaging Data
Deep learning has been leveraged by Preetham Patnam et al. (2017) to identify behaviors that precede challenging behaviors, including meltdowns and self-injurious behavior, in children with ASD. The authors took advantage of the NVIDIA Deep Learning GPU Training System (DIGITS) along with the deep learning framework Caffe (Jia et al. 2014) to rapidly train a convolutional neural network (CNN). As the name suggests, CNNs have “convolutional” and subsampling layers, allowing the model to extract features while preserving spatial relationships (Bishop 2006). Using video and images collected from various databases and Internet sources, the authors were able to train a recurrent CNN to identify common gestures displayed before a meltdown by children with ASD. A recurrent CNN has additional weights allowing cycles within the network (Yue-Hei Ng et al. 2015). On average, the model was trained in 30 to 60 min and tested on video recording of five individuals with 92% accuracy. Since the gestures were identified in less than 5 s on average, authors also developed an alarming mechanism to alert caregivers in real time. By using this model to train a more robust model and the addition of enhanced hardware, there is great potential for helping both children with ASD and their caregivers before meltdowns occur.
Recently, a deep learning classifier has been used by Heinsfeld et al. (2018) to analyze neuroimaging data. Using the ABIDE database, the study dataset was comprised of rs-fMRI scans, T1 structural brain images, and phenotypic data from 505 individuals with ASD and 530 individuals with TD across 17 sites. Dimension reduction was performed using unsupervised feed-forward neural networks called autoencoders. The model then used supervised learning by deploying two stacked autoencoders as weights to a feed-forward neural network. The deep neural network (DNN) outperformed SVM and RF classifiers with an accuracy of 70%; however, computation time exceeded 32 h (leveraging GPU-enabled computing) while the SVM and RF models finished in less than 21 min. This study shows promise for the identification of ASD biomarkers via machine learning analysis of rs-fMRIs.
Conditional Inference Forest
A CF is a modification of a RF that uses statistical inference tests to select features when constructing trees. The input variables are evaluated for independence (to the dependent variable), and the variables with the strongest association to the response are chosen (Mayor 2015). In the ASD literature included in this review, CF has been used to analyze genetic data.
Behavioral and Neuroimaging Data
Engchuan et al. (2015) used machine learning models to analyze genes, specifically rare copy number variation (CNV), associated with ASD. The dataset was comprised of 1892 participants with ASD and 2342 controls with at least one rare CNV. Using rare CNV data and comprehensive gene annotations, four classification methods were conducted and compared. The CF model’s performance was equal or superior to the other tested classification methods. The best classifier demonstrated an AUC of 0.533, correctly categorizing 7.9% of the participants with ASD while incorrectly classifying less than 3% of the controls. Performance improved when limiting the model to participants with de novo CNVs (i.e., those occurring spontaneously as opposed to inherited from a parent) or pathogenic CNVs (i.e., those that have been previously associated with ASD). Rare genic losses were found to be more predictive than gains when analyzed alone. Finally, 20 features identified as neurally relevant were found to perform better in the model than total gene count. These findings reveal the potential for machine learning to identify CNVs contributing to ASD that may not yet be well known.
Decision Trees
Classification and regression (CART) decision tree models were first introduced by Breiman et al. (1984). The classification tree predicts the likelihood of reaching the target value along with a clear path that leads to the decision. The tree is recursively constructed by posing a sequence of logical if-then conditions (from independent variables) with the answers determining the next condition (if any). Decision stumps are single-level decision trees that provide output from only one input feature (Bishop 2006). In the reviewed ASD literature, decision trees have been used to study the relationship between genetics and symptom severity in ASD, as well as predict employer recruitment of individuals with ASD.
Genetic Data
Jiao et al. (2012) used machine learning to classify children with ASD according to symptom severity using data on genetic markers. The dataset included single nucleotide polymorphism (SNP) data for 118 children with ASD between the ages of 1.5 to 14 years old. Using the results of the Childhood Autism Rating Scale (CARS; Schopler et al. 1980), participants were divided into two groups based on symptom severity. A total of 65 participants made up the mild/moderate group and 53 participants made up the severe group. Of the machine learning models evaluated, decision stumps and FlexTrees were found to perform best with an accuracy of 67%, sensitivity of 88%, and specificity of 42%. FlexTrees extend the binary-tree approach, where each split is determined by a combination of the features of interest. This suggests that SNPs have predictive power to accurately classify ASD symptom severity. One SNP in particular was found to be related to symptom severity across all the models evaluated. Results reveal that SNPs have at least moderate predictive power to make classifications based on ASD symptom severity.
Employer Survey
Hyde et al. (2018) utilized a decision tree to predict a path for individuals with ASD to successfully find employment. The model was built using 17 independent variables created from the responses of 154 representatives of various employers who have hired an individual with ASD in the past and 142 from those who have never hired an individual with ASD. The model was able to predict whether an employer has hired an individual with ASD with 75% accuracy and 82% specificity as well as identify some important features that lead to the decision.
Random Tree
Random tree classifiers use a subset of features to build a single decision tree (Mayor 2015). Like RF, random tree has been used in the reviewed ASD research to make classifications based on neuroimaging data.
Behavioral and Neuroimaging Data
Zhou et al. (2014) applied machine learning techniques to detect ASD based on multiparametric MRI data. The dataset included 127 children with ASD and 153 children with TD matched on age and gender. Cross-validation was employed in order to minimize classification error estimation bias. A model of 22 quantitative imaging features was used to predict ASD. Using a random tree classifier, a combination of four features was found to differentiate participants with ASD from those with TD with an accuracy of 68%. When six imaging features were used in the classifier, accuracy increased to 70%. Specific imaging features were also found to predict outcomes of the participants with ASD on various measures, including the ADOS, ADI-R, SRS, VASB, and IQ. These findings suggest that machine learning analyses of multiparametric MRI data may reveal biomarkers useful in the identification of ASD. Furthermore, since specific features were correlated with outcomes of behavioral measurements, such features may also prove useful in monitoring symptoms over time.
Supervised Machine Learning in ASD Text Mining
Text mining is the process of analyzing collections of texts with the aim of discovering “hidden” patterns. The Internet provides researchers practically unfettered access to current publications, but given the abundance of literature, it is nearly impossible to manually keep up to date in the field. Text mining tools allow new ways to find, sort, and discover knowledge from existing literature.
Many of the articles reviewed below applied a number of different supervised machine learning methods towards the goal of text mining. Of the 10 articles discussed, with a number of studies reporting success with more than one model, SVM algorithms were successfully used in four, Naïve Bayes was used in three, RF was used in two, Bayesian network was used in one, LASSO was used in one, finite state machine transfer network (FSMTN) was used in one, Apriori algorithm was used in one, linear variance analysis (LVA) was used in one, random walk was used in one, and ridge regression was used in one. Each of these techniques will be discussed in turn. Table 2 provides a summary of the reviewed text mining articles and the supervised machine learning methods applied.
Bayesian Network
Bayesian networks are probabilistic directed graphical models where nodes represent random variables with a corresponding conditional distribution. The arrows leaving a node point to the variable on which the distribution is conditioned. These models provide a compact representation of joint probability distributions and are a useful tool when probabilistically inferring posterior distributions or information, given new observation data (Bishop 2006). Bayesian networks have been used to resolve inconsistencies in research on molecular pathways.
Genetic Data
Rzhetsky et al. (2006) used Bayesian network to reconcile inconsistencies in published research on molecular pathways. The authors built a generalized Bayesian network of nodes and arcs from random variables to automate reconciliation endeavors on a large scale. Since it is not computationally efficient to explicitly find the joint probabilities for all states of all variables, the authors utilized a Gibbs sampler version of a Markov chain Monte Carlo technique to estimate the joint distribution values. They tested their method on 3161 research articles pertaining to the molecular interactions among genes possibly related to ASD, Alzheimer’s, bipolar disorder, and schizophrenia. The resulting network included 288 nodes and 353 arcs. The prior distributions were computed by examining 14,780 journal abstracts referring to the genes of interest. Approximately 10% of the published molecular interactions were found to be logically incompatible. These findings show the potential of text mining to resolve inconsistencies in published research that may be applied on a large scale across various disciplines.
Naïve Bayes
Naïve Bayes is a probabilistic classifier that utilizes Bayes theorem to make the (naïve) assumption that all input features are linearly independent and then compare the probability of each possible classification given a set of independent features (Bishop 2006). The naïve Bayes classifier is relatively simple and computationally inexpensive to train, and it can be competitive with more complex models like the SVM in areas like text categorization, making it still a preferred choice for spam filtering. Naïve Bayes has been used in the reviewed literature to leverage data from social media applications in order to learn more about the ASD community and to determine if ASD can be detected based on samples of writing.
Social Media
Text mining was applied to Twitter posts by Beykikhoshk et al. (2015) to gain knowledge about the ASD community. From an initial dataset of 11 million tweets, the authors used ASD-related keywords to identify over 5.6 million tweets that were ASD related. Of those, 3,493,742 were found to be original tweets created by 1,771,274 unique users. Both ASD and control corpora were found to obey Zipf’s law, suggesting that current text representation and analysis tools could be applied. The authors explored message length, word frequency, hashtag analysis, and part-of-speech analysis with classical statistics before investigating tweet classification. After splitting the data in half for training and testing sets, naïve Bayes, ridge logistic regression, and LASSO classification models were conducted. With the ASD-related keywords used to identify the ASD tweets eliminated from the analyses, naïve Bayes and logistic regression achieved approximately 80% accuracy, while LASSO had up to 70% accuracy. The authors suggested that analyses such as these could potentially yield valuable information from within the ASD community (e.g., views on current initiatives and legislature) that may be of interest to public health officials and policy makers.
Bennett (2015) also used classification algorithms to evaluate whether personal blogs could predict Autism Spectrum Quotient (AQ; Baron-Cohen et al. 2001) scores. The AQ is a questionnaire developed to aid in the diagnosis of ASD. Searching Wordpress.com for blogs containing self-reported AQ scores, Bennett identified 75 blogs (35 male, 40 female) posted after 2009 with each blog containing over 10,000 words. Analyses were conducted to determine if blog content could predict whether the authors’ AQ scores fell above or below the cutoff point for ASD. Significant differences in the number and type of useful features between male and female bloggers were found. The highest predictive accuracy, 90.9%, was achieved for males using LIWC dimension selection and a naïve Bayes classifier. Females were classified with an 88.7% accuracy using 3-g models with naïve Bayes. These finding show the potential in the use of machine learning to detect ASD from samples of text.
Finite State Machine Transfer Network
FSMTN is a transfer learning model. A transfer learning approach utilizes knowledge gained from an input source to help solve a target problem (Aggarwal and Zhai 2012). FSMTN has been used in the reviewed ASD literature to explore ASD candidate genes.
Genetic Data
Gong et al. (2011) developed the text mining tool AutMiner to explore ASD-related genes. Using PubMed’s E-Utilities, the authors created a dataset of 9276 abstracts related to ASD. An FSMTN was used to identify candidate genes, and biomedical ontologies were used to filter the results. A total of 691 genes were identified as potentially related to ASD. To validate the results, the identified genes were compared to 161 known ASD susceptibility genes. Of the 691 candidate genes, 127 genes matched known ASD susceptibility genes with 34 known susceptibility genes not appearing among the identified candidate genes. The ASD-related genes were stored in the AutMiner database, and ASD–gene and gene–gene networks were constructed. The authors proposed that AutMiner may be used by geneticists implementing genetic screenings for individuals with ASD. These findings reveal the potential of text mining in identifying ASD candidate genes.
Apriori Algorithm
Apriori algorithms look to find the k most common terms across a dataset. At each pass over the data, the algorithm adds to its prior terms other terms that appear frequently alongside the term sets it has collected previously. The Apriori algorithm is useful because it is straightforward and can be used on massive datasets relatively quickly. It generally relies on the assumption that a set cannot be frequent unless its subsets are frequent. These frequent sets are used to create association rules that expose trends in the dataset (Agrawal and Srikant 1994). It has practical uses in areas like advertising and retail sales because it is able to group items that are viewed or purchased together frequently. Like FSMTN, this algorithm has been used in the reviewed ASD literature to investigate ASD candidate genes.
Genetic Data
Gong et al. (2012) extended the work of Gong et al. (2011) using association rules. The authors extracted genes from 16,869 PubMed biomedical journal abstracts related to ASD. Based on prior knowledge, the identified genes were classified as either seed (i.e., recognized ASD susceptibility genes) or candidate. To predict new ASD susceptibility genes, association rules were created between the identified seed and candidate genes. Motivated by the Apriori algorithm, the authors developed the KF algorithm to find key factors of the candidate genes. The KF algorithm used a seed gene list, candidate gene list, and document-gene table as inputs. A total of 27 ASD susceptibility genes were found, and 43 possible susceptibility genes were predicted. These findings further underline the promising use of text mining to identify new genes potentially associated with ASD.
Support Vector Machines
SVM algorithms, described earlier in this paper, have been used in the reviewed text mining articles to reveal potentially valuable links in existing ASD research and to help protect private health information shared over the Internet.
Electronic Health Records
SVMs have also been used in text mining to help protect personal health information shared on the Internet. Song and Marsh (2012) developed a new information sharing scheme, which conceals sensitive personal information. Each patient is identified by a small set of discriminant words, which can be used to measure similarities in health conditions. The authors developed a new keyword extraction method, called linear variance analysis (LVA), which uses SVMs to calculate the variances from a centroid of the population. LVA was found to be a more effective measure of similarity than existing measures across various sets of text documents collected from online health forums, including 250 text documents with stories and experiences shared by parents of children with ASD and ADHD, 100 health descriptions related to respiratory infections, and 200 assessment reports of children being evaluated for ASD. These findings show promise in the use of text mining to help protect private health information shared over the Internet.
Ontologies
Petric et al. (2007) used text mining to uncover infrequent connections between ASD research articles in the hope of revealing new areas of exploration for researchers. A total of 214 publicly accessible articles relating to ASD were obtained from the PubMed database. The tool OntoGen (Fortuna et al. 2006) was used to create and explore various ASD ontologies. Ontologies are useful in sharing a common understanding of domain structure, explicitly formalizing domain assumptions, and analyzing domain knowledge. First, frequent research topics were explored. Using the word–vector text representation, OntoGen employed clustering to identify seven subconcepts in the ASD domain. Keywords used to describe the subconcepts were generated using a concept centroid vector and an SVM-based linear model. Rare connections between articles were also explored within the ontology. The authors specifically highlighted the relationship between ASD and calcineurin to be a potential area of future investigation. These findings demonstrate the potential of text mining to reveal infrequent and potentially valuable links within large collections of research.
Random Forest
RF, described earlier in this paper, has been used in the reviewed text mining research to explore outlier literature that link different domains.
Ontologies
Sluban et al. (2012) extended the work of Petric et al. (2007), which identified a rare connection between ASD and calcineurin (a protein that activates T cells) via text mining. Using terms that bridge ASD and calcineurin, the authors obtained 15,243 abstracts from the PubMed database. To identify sets of outlier documents that connect ASD and calcineurin, the authors employed naïve Bayes, RF, and SVM classification algorithms. The authors also compiled union and “majority” sets of outliers identified from at least two of the classification algorithms. Less than 5% of all the documents in the dataset were found to contain between 70 and 90% of the bridging terms across the five outlier subsets. Although RF was found to perform best, the authors recommended the majority set for bridging concepts. These findings reveal that text mining may be useful in filtering through literature to identify a subset of content that connects separate domains, making such research efforts less cumbersome.
ASD Monitoring
RF has also been used for ASD monitoring and surveillance. The Autism and Development Disabilities Monitoring (ADDM) Network is charged with population-based surveillance of ASD in 8-year-old children across the USA. Traditionally, clinicians manually review developmental evaluations to determine a child’s diagnostic status. Looking for a more efficient way to identify children who meet the ASD surveillance criteria, Maenner et al. (2016) trained an RF classifier on the 2008 Georgia ADDM data. A total of 5396 evaluations from 601 children with ASD and 561 children with TD were preprocessed using a bag-of-words approach with term-frequency-inverse-document-frequency (Tf-idf) weighting. RF classifiers were used to (1) identify words and phrases useful for classifying ASD and (2) classify ASD or TD. The classification model was tested on 9811 evaluations from 754 children with ASD and 696 children with TD from the 2010 Georgia ADDM data. The final RF of 3000 trees achieved 86.5% accuracy, 84% sensitivity, 89.2% specificity, and an AUC of .932. The authors note that machine learning can help differentiate children who meet the surveillance requirements; however, this model should be used more as a filter than a tool for final classification.
Random Walk
Random walk is a way to explore a space using a series of probabilistically determined steps. The probability that the next step will be in a certain direction can be predetermined. Random walks are used to calculate both expected distance after n moves as well as the probability of landing in certain regions after n steps (Bishop 2006). Random walks have been used to simulate complex behavior in many applications from sports analytics to stock prices. In the reviewed text mining literature, random walk has been used in the development of a biomedical database used to promote knowledge discovery across many disorders of interest.
Ontologies
Sulakhe et al. (2014) created Lynx, an online database and knowledge extraction tool to aid in the analysis of biomedical data. Lynx consists of an integrated knowledgebase, LynxKB, and knowledge extraction services, including the use of the text mining system GeneWays (Rzhetsky et al. 2004) and the Vista project (Visel et al. 2007). Gene prioritization was accomplished with five propagation algorithms: simple random walk, heat kernel diffusion, PageRank with priors, HITS with priors, and K-step Markov. The authors conducted a case study using Lynx to investigate the genes and molecular mechanisms associated with the occurrence of seizures in ASD. A total of 59 genes were found to be associated with ASD and seizures. After ranking the identified genes, 31 were found to be potentially related to the symptom of seizures in ASD. These findings demonstrate how text mining can be harnessed to extract knowledge from vast collections of biomedical data.
Discussion
The purpose of this paper was to review literature that has applied supervised machine learning approaches to the study of ASD. A total of 45 articles that used supervised machine learning algorithms in ASD research and text mining initiatives were identified and reviewed. In the 35 reviewed ASD research studies, the most commonly used supervised machine learning algorithms were SVM and ADtree. Supervised machine learning algorithms were used in these studies to make binary predictions (often based on diagnoses) to aid ASD diagnosis and screening efforts, explore genetic underpinnings of ASD, and identify potential ASD biomarkers (e.g., using neuroimaging). In the 10 reviewed text mining articles, the most commonly used algorithms were naïve Bayes, SVM, and Random Forest. Supervised machine learning algorithms were used in the reviewed text mining articles to explore topics of interest in online ASD communities, to identify candidate ASD genes, and to investigate obscure links between ASD and other domains.
The findings presented in this paper demonstrate that there is great value in and practical applications for the use of supervised machine learning in ASD research. Large and rich datasets, including genetic datasets, such as Krishnan et al. (2016), benefit greatly from the more flexible machine learning methods presented in this paper because these methods allow for multidimensional data to be fully utilized. Since the time from first concern to an ASD diagnosis is often over 2 years (Zwaigenbaum et al. 2009) and access to early intervention has been shown to improve outcomes (Dawson et al. 2010), the works reviewed including Duda et al. (2016b) and Abbas et al. (2018) present promising screening tools to help decrease the length of time and cost required for diagnosis. Unsupervised machine learning methods also provide practical, clinical classification tools that can be used in the field to classify or filter incoming data.
For datasets that do not have attached classification labels, supervised learning is not useful. Since outside classification can take time, money, and effort, these data can be limited. However, huge amounts of labeled data are being created in the field every day and are well fitted to this kind of supervised learning. Another limitation of machine learning comes with the complexity of the models. While the more sophisticated machine learning models often outperform their simpler counterparts, there is a tradeoff between performance and understanding. For example, the neural network presented by Linstead et al. (2017) easily outperformed a simple linear regression; however, the internal parameters of neural networks cannot be easily interpreted. Researchers should consider if this understanding is more important than the results when selecting a machine learning model.
Many of the above limitations and strengths apply to the field of Big Data and Machine Learning as a whole. Acquiring datasets large enough in terms of observations and explanatory variables to train complex models can be difficult, but with appropriately sized data, ML models often outperform simpler models. However, it is also important that these datasets are representative of the population of interest in order to avoid models that are systematically biased. While the predictive accuracy of ML models can be quite high, the models themselves are not always easily used for inferential conclusions which may decrease the adoption of such solutions. The added complexity of a ML model leads to an increase computation necessary to deploy in practical settings. This is where the science of Big Data and ML need to have the appropriate computational platform capable of handling both the data and the additional complexity of a ML model. Further research applying supervised machine learning methods to ASD research is needed. The reviewed literature has primarily applied machine learning to identify potential genetic markers and diagnosis; however, with estimates of 2% of the children diagnosed with ASD in the US (Zablotsky et al. 2015), in addition to expanding on the abovementioned areas of investigation, researchers should also investigate treatment methods, intensity, duration, and outcomes.
Availability and affordability of data collecting devices have opened doors for the use of machine learning in ASD research. In the last decade, a growing number of supervised machine learning research studies have emerged in the field of ASD. Although limited, the existing literature described in the current review has demonstrated the potential of machine learning in ASD research. The capability of machine learning to extract knowledge from large collections of data is promising for future ASD research endeavors.
References
References marked with an asterisk indicate studies included in the review
*Abbas, H., Garberson, F., Glover, E., & Wall, D. P. (2018). Machine learning approach for early detection of autism by combining questionnaire and home video screening. Journal of the American Medical Informatics Association, 25(8), 1000–1007. https://doi.org/10.1093/jamia/ocy039.
Achenbach, T. M., & Rescorla, L. (2001). Manual for the ASEBA school-age forms & profiles: an integrated system of multi-informant assessment. Burlington, VT: Research Center for Children, Youth, and Families.
Aggarwal, C. C., & Zhai, C. (Eds.). (2012). Mining text data. London, England: Springer Science & Business Media.
Agrawal, R., & Srikant, R. (1994). Fast algorithms for mining association rules. Proceedings of the 20th international conference on very large data bases (VLDB), 1215, 487–499.
American Psychiatric Association. (2013). Diagnostic and statistical manual of mental disorders (5th ed.). Arlington, VA: American Psychiatric Publishing.
Autism and Developmental Disabilities Monitoring Network. (2016). Prevalence and characteristics of autism spectrum disorder among children aged 8 years—Autism and Developmental Disabilities Monitoring Network, 11 sites, United States, 2012. Morbidity and Mortality Weekly Report Surveillance Summaries, 65, 1–23. https://doi.org/10.15585/mmwr.ss6503a1.
Baron-Cohen, S., Wheelwright, S., Skinner, R., Martin, J., & Clubley, E. (2001). The Autism-Spectrum Quotient (AQ): evidence from Asperger syndrome/high-functioning autism, males and females, scientists and mathematicians. Journal of Autism and Developmental Disorders, 31(1), 5–17. https://doi.org/10.1023/A:1005653411471.
*Bennett, S. (2015). Indicators of autism in social media: predicting autism quotient score from personal blogs (Master’s thesis). University of Edinburgh, Edinburgh.
*Beykikhoshk, A., Arandjelovic, O., Phung, D., Venkatesh, S., & Caelli, T. (2015). Using Twitter to learn about the autism community. Social Network Analysis and Mining, 5(22), 1–17. https://doi.org/10.1007/s13278-015-0261-5.
Bishop, C. M. (2006). Pattern recognition and machine learning. New York, NY: Springer.
*Bishop-Fitzpatrick, L., et al. (2018). Using machine learning to identify patterns of lifetime health problems in decedents with autism spectrum disorder. Autism Research, 11(8),1120–1128. https://doi.org/10.1002/aur.1960.
*Bone, D., Goodwin, M. S., Black, M. P., Lee, C. C., Audhkhasi, K., & Narayanan, S. (2015). Applying machine learning to facilitate autism diagnostics: pitfalls and promises. Journal of Autism and Development Disorders, 45(5), 1121–1136. https://doi.org/10.1007/s10803-014-2268-6.
*Bone, D., Bishop, S., Black, M. P., Goodwin, M. S., Lord, C., & Narayanan, S. S. (2016). Use of machine learning to improve autism screening and diagnostic instruments: effectiveness, efficiency, and multi-instrument fusion. Journal of Child Psychology and Psychiatry, 57(8), 927–937. https://doi.org/10.1111/jcpp.12559.
Breiman, L., Friedman, J., Stone, C. J., & Olshen, R. A. (1984). Classification and regression trees. Monterey, CA: Wadsworth & Brooks.
Bryson, S. E., Zwaigenbaum, L., McDermott, C., Rombough, V., & Brian, J. (2008). The Autism Observation Scale for Infants: scale development and reliability data. Journal of Autism and Developmental Disorders, 38, 731–738.
*Bussu, G., E.J. Jones, T. Charman, M.H. Johnson, J. Buitelaar, B. Team (2018). Prediction of autism at 3 years from behavioural and developmental measures in high-risk infants: a longitudinal cross-domain classifier analysis. Journal of Autism and Developmental Disorders 48, 2418–2433.
*Chen, C. P., Keown, C. L., Jahedi, A., Nair, A., Pflieger, M. E., Bailey, B. A., et al. (2015). Diagnostic classification of intrinsic functional connectivity highlights somatosensory, default mode, and visual regions in autism. NeuroImage: Clinical, 8, 238–245. https://doi.org/10.1016/j.nicl.2015.04.002.
Constantino, J. N., & Gruber, C. P. (2007). Social Responsiveness Scale (SRS). Los Angeles, CA: Western Psychological Services.
*Crippa, A., Salvatore, C., Perego, P., Forti, S., Nobile, M., Molteni, M., et al. (2015). Use of machine learning to identify children with autism and their motor abnormalities. Journal of Autism and Developmental Disorders, 45(7), 2146–2156. https://doi.org/10.1007/s10803-015-2379-8.
Dawson, G., Rogers, S., Munson, J., Smith, M., Winter, J., Greenson, J., et al. (2010). Randomized, controlled trial of an intervention for toddlers with autism: the Early Start Denver Model. Pediatrics, 125(1), e17–e23. https://doi.org/10.1542/peds.2009-0958.
Di Martino, A., Yan, C. G., Li, Q., Denio, E., Castellanos, F. X., Alaerts, K., et al. (2014). The Autism Brain Imaging Data Exchange: towards large-scale evaluation of the intrinsic brain architecture in autism. Molecular Psychiatry, 19(6), 659–667. https://doi.org/10.1038/mp.2013.78.
*Duda, M., Kosmicki, J. A., & Wall, D. P. (2014). Testing the accuracy of an observation-based classifier for rapid detection of autism risk. Translational Psychiatry, 4, 1–6. https://doi.org/10.1038/tp.2014.65.
*Duda, M., Daniels, J., & Wall, D. P. (2016a). Clinical evaluation of a novel and mobile autism risk assessment. Journal of Autism and Developmental Disorders, 46(6), 1953–1961. https://doi.org/10.1007/s10803-016-2718-4.
*Duda, M., Ma, R., Haber, N., & Wall, D. P. (2016b). Use of machine learning for behavioral distinction of autism and ADHD. Translational Psychiatry, 6, 1–5. https://doi.org/10.1038/tp.2015.221.
*Duda, M., Haber, N., Daniels, J., & Wall, D. P. (2017). Crowdsourced validation of a machine-learning classification system for autism and ADHD. Translational Psychiatry, 7(5), 1–7. https://doi.org/10.1038/tp.2017.86.
Einfeld, S. L., & Tonge, B. J. (1995). The Developmental Behaviour Checklist: the development and validation of an instrument to assess behavioural and emotional disturbance in children and adolescents with mental retardation. Journal of Autism and Developmental Disabilities, 25(2), 81–104. https://doi.org/10.1007/BF02178498.
*Engchuan, W., Dhindsa, K., Lionel, A. C., Scherer, S. W., Chan, J. H., & Merico, D. (2015). Performance of case-control rare copy number variation annotation in classification of autism. BMC Medical Genomics, 8(Suppl 1), 1–10. https://doi.org/10.1186/1755-8794-8-S1-S7.
*Florio, T., Einfeld, S., Tonge, B., & Brereton, A. (2009). Providing an independent second opinion for the diagnosis of autism using artificial intelligence over the internet. Counselling, Psychotherapy, and Health, 5(1), 232–248.
Fortuna, B., Grobelnik, M., & Mladenić, D. (2006). Semi-automatic data-driven ontology construction system. Demo at ESWC 2006, Budva, Montenegro.
Geschwind, D. H., Sowinski, J., Lord, C., Iversen, P., Shestack, J., Jones, P., et al. (2001). The Autism Genetic Resource Exchange: a resource for the study of autism and related neuropsychiatric conditions. American Journal of Human Genetics, 69(2), 463–466. https://doi.org/10.1086/321292.
*Gong, L., Sun, X., Jiang, D., & Gong, S. (2011). AutMiner: a system for extracting ASD-related genes using text mining. Journal of Biological Systems, 19(1), 113–125. https://doi.org/10.1142/S0218339011003828.
*Gong, L., Yan, Y., Xie, J., Liu, H., & Sun, X. (2012). Prediction of autism susceptibility genes based on association rules. Journal of Neuroscience Research, 90(6), 119–1125. https://doi.org/10.1002/jnr.23015.
Gotham, K., Risi, S., Pickles, A., & Lord, C. (2007). The Autism Diagnostic Observation Schedule: revised algorithms for improved diagnostic validity. Journal of Autism and Developmental Disorders, 37(4), 613–627. https://doi.org/10.1007/s10803-006-0280-1.
*Grether, J. K., Anderson, M. C., Croen, L. A., Smith, D., & Windham, G. C. (2009). Risk of autism and increasing maternal and paternal age in a large North American population. American Journal of Epidemiology, 170(9), 1118–1126. https://doi.org/10.1093/aje/kwp247.
*Heinsfeld, A.S., Franco, A. R., Craddock, C., Buchweitz, A., & Meneguzzi, F. (2018). Identification of autism spectrum disorder using deep learning and the ABIDE dataset. NeuroImage: Clinical, 17, 16–23. https://doi.org/10.1016/j.nicl.2017.08.017.
*Hyde, K., A.-J. Griffiths, C.M. Giannantonio, A.E. Hurley-Hanson, E. Linstead (2018). Predicting employer recruitment of individuals with autism spectrum disorders with decision trees. In: 17th IEEE International Conference on Machine Learning and Applications (ICMLA), Orlando, FL, 2018. IEEE.
Jia, Y., Shelhamer, E., Donahue, J., Karayev, S., Long, J., Girshick, R., et al. (2014). Caffe: convolutional architecture for fast feature embedding. Proceedings of the 22nd ACM International Conference on Multimedia, 675–678. https://doi.org/10.1145/2647868.2654889.
*Jiao, Y., Chen, R., Ke, X., Cheng, L., Chu, K., Lu, Z., et al. (2012). Single nucleotide polymorphisms predict symptom severity of autism spectrum disorder. Journal of Autism and Developmental Disorders, 42(6), 971–983. https://doi.org/10.1007/s10803-011-1327-5.
*Kosmicki, J. A., Sochat, V., Duda, M., & Wall, D. P. (2015). Searching for a minimal set of behaviors for autism detection through feature selection-based machine learning. Translational Psychiatry, 5, 1–10. https://doi.org/10.1038/tp.2015.7.
*Kou, Y., Betancur, C., Xu, H., Buxbaum, J. D., & Ma’ayan, A. (2012). Network- and attribute-based classifiers can prioritize genes and pathways for autism spectrum disorders and for intellectual disability. American Journal of Medical Genetics Part C Seminars in Medical Genetics, 160C(2), 130–142. https://doi.org/10.1002/ajmg.c.31330.
*Krishnan, A., Zang, R., Yao, V., Theesfeld, C. L., Wong, A. K., Tadych, A., et al. (2016). Genome-wide prediction and functional characterization of the genetic basis of autism spectrum disorder. Nature Neuroscience, 19(11), 1454–1462. https://doi.org/10.1038/nn.4353.
Le Couteur, A., Lord, C., & Rutter, M. (2003). Autism diagnostic interview—revised (ADI-R). Los Angeles, CA: Western Psychological Services.
*Li, B., Sharma, A., Meng, J., Purushwalkam, S., & Gowen, E. (2017). Applying machine learning to identify autistic adults using imitation: an exploratory study. PLoS One, 12(8), 1–19. https://doi.org/10.1371/journal.pone.0182652.
*Linstead, E., German, R., Dixon, D., Granpeesheh, D., Novack, M., & Powell, A. (2015). An application of neural networks to predicting mastery of learning outcomes in the treatment of autism spectrum disorder. 2015 IEEE 14th International Conference on Machine Learning and Applications (ICMLA), 414–418. https://doi.org/10.1109/ICMLA.2015.214.
*Linstead, E., Dixon, D. R., French, R., Granpeesheh, D., Adams, H., German, R., et al. (2017). Intensity and learning outcomes in the treatment of children with autism spectrum disorder. Behavior Modification, 41(2), 229–252. https://doi.org/10.1177/0145445516667059.
*Liu, W., Li, M., & Yi, L. (2016). Identifying children with autism spectrum disorder based on their face processing abnormality: a machine learning framework. Autism Research, 9(8), 888–898. https://doi.org/10.1002/aur.1615.
Lord, C., Cook, E. H., Leventhal, B. L., & Amaral, D. G. (2000a). Autism spectrum disorders. Neuron, 28(2), 355–363. https://doi.org/10.1016/S0896-6273(00)00115-X.
Lord, C., Rutter, M., DiLavore, P. C., & Risi, S. (2000b). The Autism Diagnostic Observation Schedule (ADOS). Los Angeles, CA: Western Psychological Services.
*Maenner, M. J., Yeargin-Allsopp, M., Braun, K. V. N., Christensen, D. L., & Schieve, L. A. (2016). Development of a machine learning algorithm for the surveillance of autism spectrum disorder. PLoS One, 11(12), 1–11. https://doi.org/10.1371/journal.pone.0168224.
Mayor, E. (2015). Learning predictive analytics with R. Birmingham: Packt Publishing.
Moher, D., Liberati, A., Tetzlaff, J., & Altman, D. G. (2009). Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Annals of Internal Medicine, 151(4), 264–269.
*Moradi, E., Khundrakpam, B., Lewis, J. D., Evans, A. C., & Tohka, J. (2017). Predicting symptom severity in autism spectrum disorder based on cortical thickness measures in agglomerative data. NeuroImage 144, 128–141. https://doi.org/10.1016/j.neuroimage.2016.09.049.
Mullen, E. M. (1995). Mullen scales of early learning. MN: AGS Circle Pines.
*Nakai, Y., Takiguchi, T., Matsui, G., Yamaoka, N., & Takada, S. (2017). Detecting abnormal voice prosody through single-word utterances in children with autism spectrum disorders: machine-learning-based voice analysis versus speech therapists. Perceptual and Motor Skills, 124(5), 961–973. https://doi.org/10.1177/0031512517716855.
*Petric, I., Urbancic, T., & Cestnik, B. (2007). Discovering hidden knowledge from biomedical literature. Informatica, 31(1), 15–20.
*Plitt, M., Barnes, K. A., & Martin, A. (2015). Functional connectivity classification of autism identifies highly predictive brain features but falls short of biomarker standards. NeuroImage: Clinical, 7, 359–366. https://doi.org/10.1016/j.nicl.2014.12.013.
*Preetham Patnam, V. S., George, F. T., George, K., & Verma, A. (2017). Deep learning based recognition of meltdowns in autistic kids. 2017 IEEE International Conference on Healthcare Informatics (ICHI), 391–396. https://doi.org/10.1109/ICHI.2017.35.
*Rane, S., Jolly, E., Park, A., Jang, H., & Craddock, C. (2017). Developing predictive imaging biomarkers using whole-brain classifiers: application to the ABIDE I dataset. Research Ideas and Outcomes, 3, 1–5. https://doi.org/10.3897/rio.3.e12733.
Robins, D. L., Casagrande, K., Barton, M., Chen, C. M. A., Dumont-Mathieu, T., & Fein, D. (2014). Validation of the Modified Checklist for Autism in Toddlers, Revised with Follow-Up (M-CHAT-R/F). Pediatrics, 133(1), 37–45. https://doi.org/10.1542/peds.2013-1813.
Rzhetsky, A., Iossifov, I., Koike, T., Krauthammer, M., Kra, P., Morris, M., et al. (2004). GeneWays: a system for extracting, analyzing, visualizing, and integrating molecular pathway data. Journal of Biomedical Informatics, 37(1), 43–53. https://doi.org/10.1016/j.jbi.2003.10.001.
*Rzhetsky, A., Zheng, T., & Weinreb, C. (2006). Self-correcting maps of molecular pathways. PLoS One, 1(1), 1–8. https://doi.org/10.1371/journal.pone.0000061.
Schopler, E., Reichler, R. J., DeVellis, R. F., & Daly, K. (1980). Toward objective classification of childhood autism: Childhood Autism Rating Scale (CARS). Journal of Autism and Developmental Disorders, 10(1), 91–103. https://doi.org/10.1007/BF02408436.
*Sluban, B., Jursic, M., Cestnik, B., & Lavrac, N. (2012). Exploring the power of outliers for cross-domain literature mining. In M. R. Berthold (Ed.), Bisociative knowledge discovery (pp. 325–337). Heidelberg: Springer. https://doi.org/10.1007/978-3-642-31830-6_23.
*Song, I., & Marsh, N. V. (2012). Anonymous indexing of health conditions for a similarity measure. IEEE Transactions on Information Technology in Biomedicine, 16(4), 737–744. https://doi.org/10.1109/TITB.2012.2194717.
Sparrow, S., Balla, D., & Cicchetti, D. (1984). The Vineland Adaptive Behavior Scales. Circle Pines, MN: American Guidance Services.
*Sulakhe, D., Balasubramanian, S., Xie, B., Feng, B., Taylor, A., Wang, S., et al. (2014). Lynx: a database and knowledge extraction engine for integrative medicine. Nucleic Acids Research, 42, D1007-D1012. https://doi.org/10.1093/nar/gkt1166.
Visel, A., Minovitsky, S., Dubchak, I., & Pennacchio, L. A. (2007). VISTA enhancer browser—a database of tissue-specific human enhancers. Nucleic Acids Research, 35, D88–D92. https://doi.org/10.1093/nar/gkl822.
*Wall, D. P., Dally, R., Luyster, R., Jung, J. Y., & DeLuca, T. F. (2012a). Use of artificial intelligence to shorten the behavioral diagnosis of autism. PLoS One, 7(8), 1–8. https://doi.org/10.1371/journal.pone.0043855.
*Wall, D. P., Kosmicki, J., DeLuca, T. F., Harstad, E., & Fusaro, V. A. (2012b). Use of machine learning to shorten observation-based screening and diagnosis of autism. Translational Psychiatry, 2, 1–8. https://doi.org/10.1038/tp.2012.10.
Williams, G. (2011). Data mining with Rattle and R: the art of excavating data for knowledge discovery. New York, NY: Springer.
*Xiong, H. Y., Alipanahi, B., Lee, L. J., Bretschneider, H., Merico, D., Yuen, R. K. C., et al. (2015). The human splicing code reveals new insights into the genetic determinants of disease. Science, 347(6218), 1–20. https://doi.org/10.1126/science.1254806.
*Yamagata, B. et al. (2018). Machine learning approach to identify resting-state functional connectivity pattern serving as an endophenotype of autism spectrum disorder. Brain Imaging and Behavior. https://doi.org/10.1007/s11682-018-9973-2.
Yue-Hei Ng, J., Hausknecht, M., Vijayanarasimhan, S., Vinyals, O., Monga, R., & Toderici, G. (2015). Beyond short snippets: deep networks for video classification. 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 4694–4702. https://doi.org/10.1109/CVPR.2015.7299101.
Zablotsky, B., Black, L. I., Maenner, M. J., Schieve, L. A., & Blumberg, S. J. (2015). Estimated prevalence of autism and other developmental disabilities following questionnaire changes in the 2014 National Health Interview Survey. National Health Statistics Reports, 87, 1–20.
*Zhang, F., et al. (2018). Whole brain white matter connectivity analysis using machine learning: an application to autism. Neuroimage 172, 826–837.
*Zhou, Y., Yu, F., & Duong, T. (2014). Multiparametric MRI characterization and prediction in autism spectrum disorder using graph theory and machine learning. PLoS One, 9(6), 1–10. https://doi.org/10.1371/journal.pone.0090405.
Zou, H., & Hastie, T. (2005). Regularization and variable selection via the elastic net. Journal of the Royal Statistical Society: Series B (Statistical Methodology), 67(2), 301–320. https://doi.org/10.1111/j.1467-9868.2005.00503.x.
Zwaigenbaum, L., Bryson, S., Lord, C., Rogers, S., Carter, A., Carver, L., et al. (2009). Clinical assessment and management of toddlers with suspected autism spectrum disorder: insights from studies of high-risk infants. Pediatrics, 123(5), 1383–1391. https://doi.org/10.1542/peds.2008-1606.
Funding
CPP is supported by the National Science Foundataion Division of Graduate Education, award no. #1849569.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix
Appendix

Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Hyde, K., Novack, M.N., LaHaye, N. et al. Applications of Supervised Machine Learning in Autism Spectrum Disorder Research: a Review. Rev J Autism Dev Disord 6, 128–146 (2019). https://doi.org/10.1007/s40489-019-00158-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40489-019-00158-x