Abstract
Purpose
Medical imaging is a novel research area in the domain of image processing for the research community. Features computed from MRI images provide a high level of information used in medical diagnostics. This paper addresses the classification of different types of brain tumors studied in MRI images using feature extraction techniques. It may help in the effectiveness of brain tumor treatment that depends on the early detection needed to distinguish between benign and malignant tumors.
Method
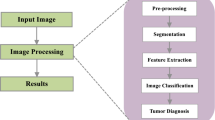
We present in this paper, a novel framework to investigate and classify brain tumors in DICOM format T2-FLAIR MRI images. Spatial filters are used to remove undesired information and noises. Segmentation is done using a thresholding method to separate the tumorous regions from healthy regions. Then, the Discrete Wavelet Transform is employed for reducing the dimensionality of the images followed by Principal Component Analysis, which reduces the dimensions further while keeping the only useful information. The gray level co-occurrence matrix is implemented to extract the texture features that can be fed into a Support Vector machine for further classification.
Results
The proposed framework is able to distinguish benign and malignant tumors precisely. The extracted features show that the benign tumor is more homogeneous with higher energy than malignant and have lower values of contrast, correlation, and entropy than malignant. The results also illustrate that benign tumors have more irregular appearance than malignant tumors. The accuracy of the proposed method is 95%. The errors in the segmentation and high dimensionality of the DICOM images causes some lack of accuracy.
Conclusion
The extracted features demonstrate that the proposed method can be used to investigate and classify the types of brain tumors. In future work, it can also be tested for tumor detection of other parts of the body. The proposed framework can be used as a quick guidance tool for the radiologists.











Similar content being viewed by others
References
Stachera, M., & Sznajder, K. (2014). Magnetic resonance in human brain examinations: A brief outline of the techniques. Utrecht: Cornell University.
Guerin, J. B., Eckel, L. J., & Kaufmann, T. J. (2009). Updates to the WHO brain tumor classification system : What the radiologist needs to know. BMC Medical Imaging. https://doi.org/10.1148/rg.2017170037.
Kurupl, M. (2015). An innovative approach to monitor brain tumor propagation and track the efficacy of treatment by processing MR Images. In IEEE (pp. 897–901). https://ieeexplore.ieee.org/document/7150870.
Shah, N. J., & Mo, K. (2012). Introduction to the basics of magnetic resonance imaging. Molecular Imaging in the Clinical Neurosciences (Springer Journal). https://doi.org/10.1007/7657_2012_56.
Aja-fern, S., & Trist, A. (2013), A review on statistical noise models for Magnetic. pp. 1–23. https://pdfs.semanticscholar.org/b67c/196652a722aa713c1619610b09b52e2cb9bf.pdf?_ga=2.175611042.105419868.1579676111-509279023.1550302710.
Abdulbaqi, H. S., & Omar, A. F. (2014), Detecting brain tumor in magnetic resonance images using hidden markov random fields and threshold techniques, In 2014 IEEE Student Conference on Research and Development (pp. 1–5). https://ieeexplore.ieee.org/document/7072963.
Singh, A. (2015). Detection of brain tumor in MRI images using combination of fuzzy C-means and SVM. In 2015 2nd International Conference on Signal Processing and Integrated Networks. (pp. 98–102). https://ieeexplore.ieee.org/document/7095308.
Maalinii, G. B., & Jatti, A. (2018). Brain tumour extraction using morphological reconstruction and thresholding. Materials Today Proceedings,5(4), 10689–10696. https://doi.org/10.1016/j.matpr.2017.12.350.
Shakeel, P. M., Tobely, T. E. E. L., Al-feel, H., Manogaran, G., & Baskar, S. (2019). Neural network based brain tumor detection using wireless infrared imaging sensor. In IEEE Access, (Vol. 7, pp. 5577–5588). https://ieeexplore.ieee.org/document/8599180.
Tiwari, A., Srivastava, S., & Pant, M. (2019). Brain tumor segmentation and classification from magnetic resonance images: review of selected methods from 2014 to 2019. Pattern Recognition Letters. https://doi.org/10.1016/j.patrec.2019.11.020.
Jemimma, T. A., & Vetharaj, Y. J. (2018). Watershed algorithm based DAPP features for brain tumor segmentation and classification. In 2018 International Conference on Smart Systems and Inventive Technology (pp. 155–158). https://ieeexplore.ieee.org/document/8748436.
Yin, B., Wang, C., & Abza, F. (2019). Biomedical signal processing and control new brain tumor classification method based on an improved version of whale optimization algorithm. Biomedical Signal Processing and Control,56, 101728. https://doi.org/10.1016/j.bspc.2019.101728.
M. Gurbină, M. Lascu, and D. Lascu (2019). Tumor detection and classification of MRI brain image using different wavelet transforms and support vector machines. (pp. 505–508). https://ieeexplore.ieee.org/abstract/document/8769040.
Huang, M., Yang, W., Wu, Y., Jiang, J., Chen, W., & Feng, Q. (2014). Brain tumor segmentation based on local independent projection-based classification. IEEE Transactions on Biomedical Engineering,61, 2633–2645.
Lindig, T., Kotikalapudi, R., Schweikardt, D., & Martin, P. (2017). Evaluation of multimodal segmentation based on 3D T1-, T2- and FLAIR- weighted images—the difficulty of choosing NeuroImage Evaluation of multimodal segmentation based on 3D T1-, T2- and FLAIR- weighted images—the difficulty of choosing. Neuroimage,170, 1–12. https://doi.org/10.1016/j.neuroimage.2017.02.016.
Talukder, K. H. & Harada, K. (2007). Discrete wavelet transform for image compression and a model of parallel image compression scheme for formal verification. In Proceedings of the World Congress on Engineering (Vol. I, pp. 2–6). https://pdfs.semanticscholar.org/38bc/52098c3e2c5517ce3dfe2dc813757dcc1f4e.pdf?_ga=2.109459845.105419868.1579676111-509279023.1550302710.
Saegusa, R., & Hashimoto, S. (2005). Pattern recognition using a nonlinear PCA 1 introduction 2 classi cation with NLPCA. In IGST (pp. 19–21). https://pdfs.semanticscholar.org/40db/8f952e267438bf9576dff716ea1dfb03e6cc.pdf.
Jimenez-del-Toro, O., Otálora, S., Andersson, M., Eurén, K., Hedlund, M., Rousson, M., Müller, H, et al. (2017). Chapter 10-analysis of histopathology images: From traditional machine learning to deep learning. In Biomedical Texture Analysis. (pp. 281–314). https://doi.org/10.1016/B978-0-12-812133-7.00010-7.
Julliand, T., Nozick, V., & Talbot, H. (2015). Image noise and digital image forensics. International Workshop on Digital Watermarking. https://doi.org/10.1007/978-3-319-31960-5_1.
Gravel, P., Beaudoin, G., & De Guise, J. A. (2004). A method for modeling noise in medical images. In IEEE Transactions on Medical Imaging (Vol. 23). https://ieeexplore.ieee.org/document/1339429.
Baselice, F., Ferraioli, G., Pascazio, V., & Sorriso, A. (2017). Bayesian MRI denoising in complex domain. Magnetic Resonance Imaging,38, 112–122. https://doi.org/10.1016/j.mri.2016.12.024.
Sharma, M. (2014). A modified and improved method for detection of tumor in brain cancer. International Journal of Computer Applications,91(6), 5–8.
Zaitoun, N. M., & Aqel, M. J. (2015). Survey on image segmentation techniques. Procedia Computer Science,65, 797–806. https://doi.org/10.1016/j.procs.2015.09.027.
Aslam, A., Khan, E., & Beg, M. M. S. (2015). Improved edge detection algorithm for brain tumor segmentation. Procedia Computer Science,58, 430–437. https://doi.org/10.1016/j.procs.2015.08.057.
Öztürk, Ş., & Akdemir, B. (2018). Application of feature extraction and classification methods for histopathological image using GLCM, LBP, LBGLCM, GLRLM and SFTA. Procedia Computer Science,132, 40–46. https://doi.org/10.1016/j.procs.2018.05.057.
Boukellouz, W., & Moussaoui, A. (2019). Magnetic resonance-driven pseudo-CT image using patch-based multi-modal feature extraction and ensemble learning with stacked generalisation. Journal of King Saud University-Computer and Information Science. https://doi.org/10.1016/j.jksuci.2019.06.002.
Yadav, J., & Sehra, K. (2018). Scale Dual l Tree Complex Wavelet Transform based robust features in PCA subspace for digital image watermarking features in PCA and SVD subspace for digital image watermarking. Procedia Computer Science,132, 863–872. https://doi.org/10.1016/j.procs.2018.05.098.
Arabi, P. M., Joshi, G., & Deepa, N. V. (2016). Performance evaluation of GLCM and pixel intensity matrix for skin texture analysis. Perspectives in Science,8, 203–206. https://doi.org/10.1016/j.pisc.2016.03.018.
Shree, N. V. (2018). Identification and classification of brain tumor MRI images with feature extraction using DWT and probabilistic neural network. Brian Informatics.,5(1), 23–30. https://doi.org/10.1007/s40708-017-0075-5.pdf.
Alqazzaz, S. (2019). Automated brain tumor segmentation on multi-modal MR image using SegNet. Computational Visual Media,5(2), 209–219. https://doi.org/10.1007/s41095-019-0139-y.
Kumar, S., Dabas, C., & Godara, S. (2017). Classification of brain MRI tumor images : A hybrid approach. Procedia Computer Science,122, 510–517. https://doi.org/10.1016/j.procs.2017.11.400.
E. Sankar (2017). Digital image processing Image enhancement in spatial filtering, DST sponsored national conference held on recent trends, advancement and applications in digital image processing NCDIP-2016, At SCSVMV University, Kanchipuram. (pp. 1–3). https://www.researchgate.net/publication/312595178_Digital_image_processing_Image_Enhancement_in_Spatial_Filtering.
Bhargavi, K. (2014). A survey on threshold based segmentation technique in image processing. International Journal Corner,3(12), 234–239.
Burnase, S. R., & Swamy, S. (2014). Hyperspectral image reduction using discrete wavelet transform. Journal of Electrical and Electronics Engineering,43, 13–16.
Sahooinst, R., Roy, S., & Chaudhuri, S. S. (2014). Haar wavelet transform image compression using various run length encoding schemes. In Proceedings of the 3rd International Conference on Frontiers of Intelligent Computing: Theory and Application (pp. 37–42). Springer. https://doi.org/10.1007/978-3-319-11933-5_5.
Ng, S. C. (2017). Principal component analysis to reduce dimensions on digital image compression. Procedia Computer Science,111, 113–119. https://doi.org/10.1016/j.procs.2017.06.017.
Ali, F., El-khoribi, R. A., & Shoman, M. E. (2016). A novel brain computer interface based on principle component analysis. Procedia Computer Science,82(3), 49–56.
Al-ani, M. S., & Alheeti, K. M. A. (2017). Precision statistical analysis of images based on brightness distribution. Advances in Science, Technology and Engineering System Journal,2(4), 99–104.
Snehkunj, R., Jani, A. N., & Jani, N. N. (2018). Brain MRI/CT images feature extraction to enhance abnormalities quantification. Indian Journal of Science & Technology,11(1), 1–10.
Zayed, N., & Elnemr, H. A. (2015). Statistical analysis of Haralick texture features to discriminate lung abnormalities. International Journal of Biomedical Imaging. https://doi.org/10.1155/2015/267807.
Wang. L. (Ed.). (2005). Support vector machines: Theory and applications vector machines. Computer Science. Vol. 177, p. 6221. https://www.springer.com/gp/book/9783540243885.
Funding
This paper does not have funding.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors confirm that this article content has no conflict of interest.
Ethical Approval
This paper does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Hamid, M.A.A., Khan, N.A. Investigation and Classification of MRI Brain Tumors Using Feature Extraction Technique. J. Med. Biol. Eng. 40, 307–317 (2020). https://doi.org/10.1007/s40846-020-00510-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40846-020-00510-1