Abstract
The enormous outbreak of biomedical knowledge, the aim of reducing computation and processing costs and the widespread availability of internet connection have created a profuse amount of electronic data. Such data are stored across the globe in various data sources that are semantically, structurally and syntactically different. This decentralized nature of biomedical data has made it difficult to obtain a unified view of the data. Data integration plays a crucial role in enhancing access to heterogeneous data making the retrieval easier and faster. A variety of ontology, machine learning, deep learning and fuzzy logic-based solutions are being developed for heterogeneous data integration. The proposed model concentrates on the automatic ontology-based data integration method that can be effectively deployed and used in the healthcare domain. The proposed model is divided into three phases. The first phase includes the automatic mapping of data and generation of local ontology across heterogeneous data sources, the second phase combines the local ontology models developed in the first phase to create a root global schema mapping and the third phase queries diverse databases to retrieve semantically analogous records. The model is created based on the medical records, chest X-ray details and COVID-19 symptom questionnaire data of various patients distributed across three data sources (SQL, mongodb and excel). Based on the data, the patients who have moderate/higher risk of developing serious illness from COVID-19 are retrieved.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ontology is a method of knowledge representation that represents knowledge as a set of concepts within a domain. Ontologies typically include a shared vocabulary that explains the kind, behavior, and relationships between the domain's concepts. They’re most typically utilized to derive relevant insights from a variety of data sources. As a result, they have a wide range of applications in fields such as Artificial Intelligence, Big Data Analytics, Semantic Web, Library Management, Biomedical domain, and so on.
The digitization of patient records has resulted in the development of a large amount of data, which will continue to grow. X-rays, scans, temperature sensors, test findings, prescriptions, monitoring devices, and other sources generate these patient records; this big data has three major characteristics: diversity, velocity, and volume.
The heterogeneity found in the data supplied by the aforementioned sources is referred to as variety. The pace at which data is created, processed, analyzed, and stored across numerous platforms is referred to as velocity. The quantity of data created by the data sources is referred to as volume. In all stages of biological data management, such as data acquisition, integration, and storage, ontology is a key tool for handling huge data [1].
Phases of Ontology
Ontology engineering is divided into three phases: ontology building, ontology mapping, and ontology integration. Many manual approaches for the three phases mentioned above have been proposed. The three key phases of ontology engineering are increasingly being automated with more attention. Based on Domain Specific Word embeddings [2], one such automated ontology matching technique was developed. The semantic meaning of the entities was extracted using this method by training word vectors on the biomedical corpus. The word embedding technique was then applied to existing systems, with excellent results. The semantic issues that arise in the biological sector were handled with this strategy.
Types of Ontology
Ontologies are divided into three categories based on the application’s outlook. The first is referred to as an upper ontology [3]. This ontology supports in the determination and organization of terms and associations in a specific area by providing a basic framework.
A reference ontology, on the other hand, represents the objects and connections in a single domain. The third category is an application ontology, which can be utilized in a specific scenario and provides a basic dictionary to meet the requirements. Based on the scenario considered for development, an ontology-based solution designed to handle a heterogeneity problem can fall into one of the three categories above.
Solving Heterogeneity Problems
Data integration can be done using either data translation or query translation to tackle heterogeneity issues. When the data translation approach is utilized, all of the data in a database is translated. In the query translation approach, the query from the user application is turned into a format that can be used to obtain data from numerous data sources using appropriate software.
Query translation is the more efficient of the two strategies presented since it is quick and has less overhead. The data translation technique, on the other hand, has the drawback of taking a long time to translate data and is less efficient when new data is introduced. After then, the data must be translated to match the established rule before being accessible. The results of an ontology model can be effectively obtained using SPARQL, an ontology-based query language.
Issues in Data Integration
Data consolidation is another difficult topic in the field of data integration. When many data sources are combined, the data is more likely to be redundant, transitively dependent, and have multi-valued properties. As a result, prior to ontology building and mapping, the data must be normalized, as unnormalized data can lead to less efficient results.
Normalization can be used to three sorts of granularities in data integration: record-level, field-level, and value-level components. Many academics have surveyed the key integrity concerns encountered in the biomedical area during heterogeneous data integration and have proposed feasible solutions. The study covers the most important data integrity approach utilized in the biomedical domain and discusses data integrity strategies employed in that domain. In the biomedical industry, an extensive systematic literature review is conducted. The main purpose of the systematic literature review (SLR) [4] is to find out what data integrity strategies are currently being used by researchers in the biomedical area.
This SLR was done in two parts. The SLR gives thorough information regarding previous data integrity threats linked to the biomedical industry in the first stage. This section discusses data breaches in various healthcare industries.
The SLR presents a thorough analysis of historical research endeavors related to biological data integrity that aid in the security of healthcare systems in the second stage. Only standard articles in the biomedical sector were examined for inclusion in the study.
Other Data Integration Techniques
Various integration solutions are studied, including blockchain and masked authenticated messaging extension, and the optimum integrity solution for each healthcare domain is offered. According to the study, secure authentication-based access can be used in healthcare systems. For data sharing, blockchain, secure cloud, and Slepian wolf coding based secret sharing may be used; for patient data access, cryptography and Merkle tree based approach can be used; and for data storage, secure cloud and blockchain can be used.
A privacy-free data fusion and mining (PFDM) technique was used to combine data in the healthcare sector in a time-efficient and privacy-preserving way.
The major contribution of the model was separated into three components.
-
i)
The LSH (locality-sensitive hashing) was created for multi-source IoH (Internet of Health) data fusion and integration to safeguard critical patient information that was not explicitly available in earlier IoH data.
-
ii)
After the LSH technique, an analogous IoH data record search strategy for future IoH data mining and canvas is proposed for Internet of Health data without any sensitive patient information.
-
iii)
A series of pre-designed tests based on data collected from real-world customers confirms the advantages of the PFDM work.
First, LSH functions are used to project sensitive IoH information. Then, using the IoH data record and the matching hash values produced during hash projection, a collection of hash tables is created. Finally, comparable IoH data mining and search are performed using the traced hash tables.
User based collaborative filtering (UCF) and item based collaborative filtering PDFM were used to compare this method. LSH returned IoH data records in a short amount of time, with a lower mean absolute error rate and a higher similarity index. Only a simple IoH data of continuous type is studied, with data type diversity (e.g., continuous data, discrete data, Boolean data) and data structure diversity (e.g., continuous data, discrete data, Boolean data) not taken into account.
PDFM [5] does not yet have the capability of safeguarding sensitive patient information. Data privacy and data availability are frequently inextricably linked. It is not always possible to ensure data availability. By taking into account the potential diversity of data types and data structure, the suggested PDFM approach can be upgraded. Combining numerous existing privacy solutions for improved performance is still a work in progress that requires ongoing research. Ontologies can be effectively used to secure patient information via security recommendation systems.
Based on the requirements, data is stored in diverse data sources. Relational database is preferred when there is a rigid schema for the entities and numerous relations exist between them. Document databases are preferred when the data to be stored is semi-structured and flexible. In case of questionnaires, excel data source is taken into consideration. The proposed method aims at integrating the data stored across the aforementioned data sources, i.e., relational, document-based and excel data sources using ontology-based techniques and querying them to monitor the COVID-19 symptoms in a patient.
The relational data source used in the proposed system is SQL Server and it holds the patient medical records. MongoDB is the document-based data source taken into consideration and description of chest X-ray details are stored in it. The answers for symptom questionnaire are stored in Microsoft excel data source.
The paper is divided into six sections. The first section provides a brief introduction on all the ontology-based techniques and types used in various domains. The second section includes the related works in data integration domain using ontology. The third section describes the architecture of the proposed model, the methods of constructing local and global schemas, and querying them. The implementation screenshots are presented in the fourth section followed by results and discussions in the fifth section. The sixth section describes the conclusion and future work of the proposed system.
Related Work
Relational Models
Considerable number of studies have been conducted on building ontologies from relational databases. The method proposed by Asfand-e-yar and Ali [6], involves building an ontology model for a relational database and a flat file database. The models were then combined to form a generic model. In the last phase, the data was queried using SPARQL to retrieve the book details from the library data. The model was simple and efficient but lacked automatic methods of mapping and integration.
Another approach by Li et al. [7], introduced a novel model of data integration for monitoring the bridge conditions based on sensor data stored in MySQL databases. This model was labeled as structural health monitoring systems and was implemented on an actual bridge SHM big data platform in Ningxia, China (taking 2 bridges A and B). Implementation of sensory data integration was based on the R2RML engine. Furthermore, the querying and reasoning engines are built using SPARQL, RDF schema, OWL, and SWRL semantics. The SWRL rule gets the maximum measurement range value. If the observed data exceeds the threshold, a fault discovered value is set to true, and the related sensor administrator is notified by sending an automated notice.
The model was more convenient and intelligent for sensory data retrieval and system management with the assistance of ontologies-based querying and reasoning. Because of the unique nature of bridge constructions, the model is limited in its ability to provide good damage forecast using just big data-driven methods. The proposed model can be further improved to handle all the activities during the whole life cycle of bridges.
Normalization Methods
When integrating data that is stored across various databases, special emphasis must be laid on removing the redundancy of data. The presence of improper data reduces the efficiency and usefulness of the process. Therefore, it is necessary to normalize the data before mapping them to ontology models. The article by Dong et al. [8] gives a brief overview of record normalization approaches, ranging from simple methods that rely solely on data from records to complicated strategies that mine a group of duplicate records before determining a value for a normalized record's attribute.
This study identifies three degrees of normalization granularity: record, field, and value component. To pick the normalized record or the normalized field value, the four single-strategy approaches: frequency, length, centroid, and feature-based were analyzed. The results from a variety of single strategies, such as borda-based approach and weighted-borda-based approach, were combined using result merging models inspired by meta searching for the multi-strategy approach.
Of all the approaches discussed, the WBorda approach outperformed all the other methodologies. For single-strategy feature-based ranker (FBR) was shown to perform the best. The experiments discussed here lacked diversity and had less manual interaction. Since automated solutions alone will not be able to reach flawless accuracy and produce solutions that can handle numeric or more complicated values, the work discussed above may be enhanced to include an effective human-in-the-loop component in the existing solution.
Tools and Technologies
Owing to the increased generation of data in the biomedical field and the need to effectively handle, retrieve and process data, the ontology-based integration and retrieval plays a very important role in the healthcare domain. The paper published by Dhanye et al. [9], contains a detailed survey on data integration technologies, tools, and applications within the healthcare domain.
The Integration Methodologies For Big Healthcare Data like Data Consolidation (Data Lake, Apache Hive, HiveQL), data virtualization (SAP HANA) and data propagation (SparkMed), Integration Technologies like Semantic Web (RDF, OWL, SPARQL, Linked Open Data) and Machine Learning (Supervised and Unsupervised Learning methods, ANN and Deep learning models.) and Integration Tools like Ingestion Tools—Flume (log data), Sqoop (transfer RDBMS data to HDFS), Apache NiFi (Diverse data source), Storage Tools—File System(Amazon S3, HDFS), NoSQL (Redis, Cassandra, CouchDB,etc.,), Data Warehouse (Hive), Processing tools—Batch Processing (Map Reduce, Spark SQL), Stream Processing (Kafka, Flink) and Machine Learning (MLib, Google’s sensor flow) technologies were discussed. Each technology along with its advantages and shortcomings were explored. However parallel programming models were not considered in this study. The work can be further extended to explore the parallel processing models that could serve as the potential solutions for integrating Big Healthcare Data.
HealthCare Domain
Peral et al. [10] suggested an ontology-oriented framework for integrating heterogeneous data from telemedicine systems. Natural language processing (NLP) and artificial intelligence (AI) approaches were utilized to integrate data from several heterogeneous sources utilizing a core ontology as a knowledge basis. The method was used in both personalized medicine (the study, diagnosis, and treatment of illnesses that are unique to each patient) and telemedicine (here diabetes). There are two steps to creating this ontology-oriented architecture: data collection and data running.
In the data collection phase, the health data was collected from web data, structured database, sensor data and other sources. The result of this phase had a set of different kinds of rules called integrated rules. The web rule extraction was done using NLP and DM techniques like document selection, information retrieval (IR) task etc.,
The information acquired from the sensors (sensor data) and the incorporated DM rules are transferred to the telemedicine system during the running phase. If the system identifies any aberrant measurements, an alarm is sent to the medical staff. The team can analyze and can either accept/reject the alarm. The approach used weka’s Time series analysis to predict glucose levels based on the historical data. SVM and linear regression were used to specify the range for the risk of diabetes. The system provided the medical team with real-time information for making the right medical decisions to prevent possible deteriorations of type 1 diabetes patients. However, this model was not that flexible framework for real-time embedded systems because of the limited handling of the Velocity aspect of Big Data. The model can be improved by including social media as a part of the dataset and using natural language processing and artificial intelligence methods to automate the process.
Another ontology-based data access method in the healthcare domain was proposed by Zhang et al. [11], brings out a case study that integrates three datasets and predicts the cancer survival rates using cox proportional hazard models. To fully understand the effect among predictors, the heterogeneous datasets are pooled for integrative data analysis (IDA). The method involved the following steps.
Step 1: Create a query interface by synthesizing and integrating descriptions from many data sources.
Step 2: As a metadata representation of the data items, a global ontology is created. The link between data sources and within them is established.
Step 3: Semantic mappings between the global ontology and the data sources are established.
Step 4: Using the semantic mappings, a high-level semantic inquiry is re-formulated into a union of sub-queries across all data sources and executed using SPARQL. The global ontology, the Ontology for Cancer Research Variables (OCRV), was built using protege, utilizing the Web Ontology Language (OWL) and the Ontop Protégé plugin.
Semantic data integration was then implemented using On-Top Protégé. The implementation methods involved construction of OCRV and building Semantic queries using RDF graphs. Finally, a data Integration pipeline was created using OWL API and SPARQL. Various queries related to cancer survival rates are executed for different use cases. The model described lacks automation as the global ontology and semantic mappings are constructed manually. The further enhancement of this work can include automatic construction of global ontology and semantic mappings.
Semi-Automatic Ontology Creation
A semi-automatic method of creating ontology by extracting the database schema was proposed by Zhao et al. [12]. The Ontology integration was done by means of relational algebra and the rooted graph techniques. SPARQL was used for making semantic queries. The two-world famous First Principle Computational databases, Open Quantum Materials Database (OQMD) and Materials Project were used as the integration targets, which show the availability and effectiveness of this method. The semantic integration was done by extracting the relational model from the database using the database connection API. Then material ontology was generated according to the relational model and conversion rules.
According to the conversion rules, the database tuples are transformed to ontology individuals. For the materials ontology and other materials database, an algebraic model is created. After that, a relationship is established between them to translate database data into ontology entities. The heterogeneous databases may be linked together using these techniques. Ontology entities are utilized as the data carrier when employing relation algebra to combine the ontology and the database, which is better ideal for SPARQL. To improve the accuracy of query results, ontology rules are used.
The query performance from the model can be further improved by physically separating the data and ontology. Visualization of SPARQL construction can be done to make it easier for normal non-professional users to use the system friendly and conveniently.
Semantic Method-Based Ontology
Hazber et al. [13] introduced a unique methodology that allows semantic web applications to use semantic approaches to access relational databases and their contents. This method has two primary phases: creating an ontology from an RDB schema and automatically generating ontology instances from RDB data. The JDBC driver engine in Java was used to extract metadata (schema) and data from an RDB. Tables, columns with data types, relationships, integrity constraints, referential constraints, and check constraints are all part of the schema analysis.
The RDB model is then transformed into the Ontology model by mapping rules. Apache Jena package and transformation functions were used to generate the output based on the OWL structure built on RDF(S). RDF triples are then generated. A generic ontology was finally developed and an ontology validator was used to verify the generated ontology. This method allowed for a direct mapping of relational database Schema to a semantic web ontology constructed in OWL on top of RDF utilizing the RDFS vocabulary and the XML Schema data type. One of the key advantages of this technique was the ability to examine various RDB scenarios, which reduced information loss and eliminated uncertainty when rules were applied.
The technique had a flaw in that it didn't examine the database's static structures. Extraction of additional mapping rules and querying relational databases on the semantic web using an ontology created by the rules can be added to the effort. Only the static architecture of relational databases is investigated, leaving dynamic components such as triggers to be investigated in the future.
Ontology for NoSQL Databases
The work proposed by Banerjee et al. [14]. took the NoSQL databases under consideration and constructed an ontology driven query language. This model provided a common representation for both schema based and schema-less representation of data. The following operators in NoSQL databases are explained in the paper.
-
i)
Check Construct-Type Operator—This operator checks whether the construct type is Family/Collection/Attribute
-
ii)
Get_Path Operators—Specify return paths from collections
-
iii)
Create Operators—Create families and collections in ontology-based query language
-
iv)
Write Operator—Write records into collections
-
v)
Select Operator—Select specific families/Attributes in collection
-
vi)
Retrieve Operator—Retrieve specific families in collection.
-
vii)
Update Operator—Modify Collections/Families
-
viii)
Delete Operator—Remove records from collection
-
ix)
Aggregate Operators—Perform Operations like sum, count, group, set union, set difference and set intersection on records
Based on the above operators, a querying system was developed and executed for an E-Prescription Collection. This Query language improved the efficiency of query execution and portability of data. However, this model lacked rule language definition for efficient query answering. The operators referred to in the paper can be transformed into native NoSQL databases. A formal semantics of query operators can further be built in a rule language for efficient query answering.
Ontology Learning Life Cycle
A novel approach proposed by Bilal Ben Mahria, et al. [15], introduces a new life cycle for ontology learning from relational databases based on software engineering requirements.
The life cycle of learning ontology from the relational database includes following phases.
(1) Discovery:
The domain and scope of the research are covered in this stage (by means of a questionnaire).
(2) Preparation:
Data cleaning, pre-processing, normalization and conditioning is carried out in this phase.
(3) Development:
Ontology building takes place here. Data Acquisition (ABox), Schema Acquisition (TBox), alignment, merging and integration are done here.
(4) Evaluation
Evaluation is made in two dimensions
-
i)
T-Box evaluation and
-
ii)
A-Box evaluation.
The generation and evaluation of T-Box and A-Box are done using OWL language and near to program development language.
The following metrics for evaluating the model are calculated
-
i)
Attribute richness (AR)
-
ii)
Relationship richness (IR)
-
iii)
Inheritance richness (RR)
-
iv)
Average population (AP)
-
v)
Class Richness (CR)
To show the efficiency of the process, a study was conducted on six relational databases from the e-commerce domain. To achieve good results, the RDB that weren’t semantically strong were removed. However, the paper focuses only on relational databases. Further studies can be done to explore the scope of this life cycle model to unstructured databases.
Ontologies in Biomedical Domain
The biomedical domain encompasses a variety of fields that use elements of natural science, formal science, or both to generate information, medicines, or technology for healthcare and public health. Biomedical sciences include medical microbiology, clinical virology, clinical epidemiology, genetic epidemiology, and biomedical engineering. The acquisition of ontological relations, the integration of diverse datasets, and biological knowledge reasoning utilizing ontology are all part of biomedical ontology research.
Because of the valuable applications provided by biomedical ontologies, biologists will be able to keep up with an ever-increasing volume of data. They will also be liberated from the less fascinating tasks that can be automated using ontologies.
Cardiology
Physical fitness is very crucial for cardio vascular disease patients. Another ontology, OPTImAL [16], was created to see if CVD patients followed their physical activity, yoga, and exercise regimens to the letter. The NeOn framework was used to modify the ontology, which was produced using the Ontology Development 101 technique. It was created with the OWL2 programming language, Protégé, WEBVOWL, and Ontograph with the OAF plugin. To signify the patient profile factors, OPTImAL has 142 classes, 10 object properties, and 371 persons. A cardiologist and three Cardiac Rehabilitation trainers assessed the model for its applicability and effectiveness.
Nephrology
To benefit researchers and nephrology practitioners, a wide assessment of ontologies in the nephrology domain was conducted. The responsibilities of ontology in biomedical sciences are discussed in this article, as well as the unavoidable role of data integration in accessing huge data. The role of Kidney Precision Medical Project (KPMP) ontologies [17] in bridging the gaps of kidney specific data representation was investigated using current ontologies in the nephrology domain. The use of the created ontologies to aid in the data harmonization of terms relevant to renal disease was investigated. The concepts of two ontology resources Ontology of Precision Medicine and Investigation (OPMI) and, Kidney Tissue Atlas Ontology (KTAO) that may be utilized to evaluate KPMP data, were given a lot of attention.
Diabetes
An Ontology Network for Diabetes was built using three key resources: the drug catalog of diabetic patients, the patient’s medical history and notes, and the previous 5 years’ epidemiological hints of type 2 diabetes mellitus (T2DM). Six ontologies [18] relating to the diabetes domain were developed using the information above, followed by Ontology reuse and integration. The network was built in RDF and further upgraded by the use of semantic web rule language (SWRL). The data was retrieved from the ontology using SPARQL queries. A few non-ontological data points, such as patient notes written in plain language, were also included in the network. This network’s future additions could include the generation of notifications for excessive glucose and cholesterol levels.
COVID-19
The Coronavirus, commonly known as COVID-19, has been declared a pandemic by the World Health Organization (WHO). More than 220 million cases have been confirmed, with 4.6 million people dying as a result. In individuals who have already been infected, this extremely infectious respiratory disease manifests itself in both symptomatic and asymptomatic patterns, resulting to an exponential increase in the number of disease contractions and fatalities. Coronavirus Infectious Disease Ontology (CIDO) [19] was developed to provide ontology-based solutions for coronavirus disease transmission, pathophysiology, epidemiology, prevention, diagnosis, and treatment. CIDO follows the Open Biomedical and Biological Ontologies (OBO) principles and appropriate ontology building methodologies. The future work of this model might be used to create and analyze vaccines to eliminate COVID-19.
Traditional Medicine
Traditional medicine includes plant, animal, and mineral-related medications, spiritual forms of treatment, home remedies, and workouts, among other health practices, techniques, knowledge, and beliefs. They are used to cure, detect, and minimize the danger of infection, as well as to maintain health, either alone or in combination.
Ayurveda, Siddha, Yoga, homeopathy, and Unani are some of the traditional medicines practiced in India. Ayurveda contains a large quantity of unstructured data, hence Ontology [20] might be useful in deciphering the information from the unstructured data. When compared to machine learning methods for similar challenges, an Ontology model based on Biomedical Text Mining (BTM) was constructed and proved to have superior accuracy in classification of medical texts. Text collection, processing, and analysis were among the most important aspects of the model’s development. The information was acquired from the traditional knowledge digital library (TKDL), and therapeutic plants such as aloe vera, ginger, turmeric, tulsi, and others were considered.
In the existing works, various methodologies were suggested to integrate data from similar kind of data source with different schema. The proposed method integrates the data from three heterogenous data sources (SQL Server, MongoDB and Excel) using XML-based global schema and queries them to retrieve the patient details.
Proposed System
All the ontology-based existing systems for data integration from heterogeneous data sources deal with integrating data from same kind of datastore (either relational/ non-relational) but with different schema. Certain integration techniques were also proposed to combine relational and non-relational data sources. The proposed system combines the data from SQL, MongoDB and Spreadsheet to detect the patients at high risk of COVID-19 symptoms as selected by the doctor. Storing data across different data source kinds helps in faster retrieval of the required data. The Proposed system helps the doctor to query the patient records stored across various data sources without the knowledge of the query required to access them.
Data Stores Used
Three different datastores are used in the proposed system. They are
-
i)
Excel
-
ii)
SQL
-
iii)
MongoDB
Excel
Microsoft Excel is a spreadsheet software that is available for a number of operating systems. Excel includes calculators, graphing tools, pivot tables, and visual basic for applications, a macro programming language. Excel based calculations are widely used for entry and storage of data, completing calculations, data interpretation and analysis, visualizations and reporting, budgeting and accounting, schedules and calendars, administrative and management responsibilities, automating repetitive tasks and forecasting.
The Excel sheet contains the symptom questionnaire details filled by the patient. The Symptom questionnaire has questions about various symptoms of COVID-19, i.e., does the patient have fever; does the patient have loss of smell/ taste etc., The patient has to answer yes/no for those questions.
SQL Server
Microsoft SQL Server is a relational database management system developed by Microsoft. It’s a database server, a software product whose primary function is to store and retrieve data for other software programs that may operate on the same computer or over a network.
The patient’s medical history stating if he/she has diabetes, cholesterol, pneumonia, etc. are stored in the SQL Server. The patient’s contact details (name, address, and contact number) are also stored in SQL. This detail helps the doctor in contacting the patient with the risk of selected COVID-19 symptoms.
MongoDB
MongoDB is a document-based database that can store a large quantity of data while allowing you to process it rapidly. MongoDB is a NoSQL (Not Only SQL) database since it stores and retrieves data in the form of documents rather than tables. MongoDB is a database that does not have a schema.
A schema-less database allows diverse documents with varied structures to be stored in the same collection. To put it another way, a single MongoDB collection may store several documents, each with a varied number of fields, content, and length. Unlike relational databases, one document does not have to be identical to another. MongoDB provides a lot of flexibility to databases.
The chest X-ray details of the patients are stored in MongoDB. Few fields were absent for most of the records and hence the schema-less representation was used to store the X-ray details. The presence of A-lines, B-line, comments from medical doctor, preconditions about the patient and other details from the chest X-ray were stored in MongoDB.
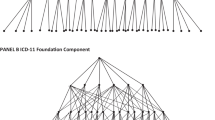
System Architecture
The system architecture of the proposed system is represented in Fig. 1 and the processes are explained below.
Local Schema
The database schema is a formal linguistic description of the database structure that is understood by the database management system (DBMS). The term “schema” relates to how data is organized and how a database is constructed (divided into database tables in the case of relational databases). Integrity constraints are a set of formulae (sentences) that are imposed on a database to formalize a database structure. These constraints guarantee that the schema’s elements are compatible. All the constraints that a datastore should satisfy are expressed in a database language. This database language can be further used to create a datastore. The local schema formed is then combined with other schemas of the datastores involved in the integration process, to form a global schema. This explains how real-world entities are represented in a database. The local schema refers to mapping from a single table/collection/sheet to a real word entity.
Three local schemas were constructed for Excel, SQL, and MongoDB in the proposed system, respectively. Java utilities were created for local schema generation, which automatically reads the data source and converts them into its respective local schema using the libraries and dependencies. Instead of storing the entire data from excel, SQL and MongoDB, only the schema is stored. Storing the schema alone saves a considerable amount of space and prevents Insert, Delete and Update anomalies.
Local Schema Generation from Excel
The local schema generation from Excel is done using Apache poi library. The Apache POI project, which was originally a sub-project of the Jakarta Project, offers pure Java libraries for reading and writing Microsoft Office files such as Word, PowerPoint, and Excel. The File object was created and the path of the csv file was given. XSSF Workbook was used to read the file into the java object. The sheet at the first index was retrieved. The first row has the attributes for the symptom questionnaire data. The row was read and was written into an XML file along with its datatype.
Local Schema Generation from SQL
Connection with SQL server was made using the MSSQL library by SQL server authentication providing the username and password. Once the connection is established, the schema can be extracted using the name and type field from the sys.columns table providing the table name as the object ID. Once the fields are retrieved, the local Schema is extracted and written into an XML File.
Local Schema Generation from MongoDB
The connection with MongoDB was established using the mongo driver core library. The connection is established by providing the port number 27017 of the system in the java utility.
Since we have only a single table/collection/sheet in the data source, the local XML schema represents our local ontology.
Global Schema Creation
A global schema is a single, connected view of heterogeneous databases. It represents the logical conceptual view of the system. The local schema created for the data sources are merged and written into an XML global schema.
Mapping File Generation
Data mapping is a critical design phase in data migration, data integration, and transformation initiatives. Artificial intelligence is used in modern systems to map data fields from a source format to a destination format. They are used to let the system know where the data resides. Data mapping is a critical component of assuring data accuracy throughout data transfer from a source to a destination. Data quality in the data warehouse is ensured by good data mapping.
The global schema created in the above step is read and the mapping file is created in XML. The mapping file has the name of the data source in tags and within the tags the attributes present in the data source are present.
The global schema and the mapping file are then parsed using a JDOM parser to facilitate the data access.
JDOM Parser
The World Wide Web Consortium’s official proposal is the Document Object Model (DOM) (W3C). It specifies an interface that allows programs to modify the style, structure, and content of XML documents. This interface is implemented by XML parsers that support DOM.
Reading the Global Schema and Mapping File Using JDOM Parser
To parse the attributes using a DOM parser, the mapping file is loaded into a file object. A new Instance for the Document Builder Factory is created. The file object is the parsed and stored into a document. A node list is created by reading all the elements between the map tag. An attribute map has the tag value and the parent tag value, where tag value is the actual attribute and the parent tag value denotes the data source in which the attribute is located.
Formulate the Query
In the proposed model query formulation has two parts
-
i)
Symptom generator query
-
ii)
Output field generator query
(1) Symptom generator query
This query is used to fetch all the fields having yes/no values from three data sources (Excel, SQL, MongoDB) and display them in the User Interface (UI). The doctor can select the required COVID-19 symptom field and get the patients, who are at high risk of the selected symptoms. The details regarding the residence of the data are obtained from the attribute map created using the DOM parser.
(a) Excel based symptom field generator
This is done based on Apache POI library. The columns present in the csv file are retrieved and the cells are checked for yes/no values using the getStringValue() function. If the entire column contains none but the two values yes/no, then it is marked as a symptom field and written into an XML file.
(b) SQL based symptom field generator
A connection with SQL Server is established and a query that extracts the columns having the values Yes/No is created. Based on query’s result set, the corresponding columns are written into the XML File.
(c) MongoDB based symptom field generator
To fetch the columns representing symptoms from a MongoDB collection, a MongoClient is created for localhost with the port number 27017. A connection is established using the database name and collection name. From the DOM parser, we get the attributes stored in MongoDB and create a projection in the collection using those fields. Then the fields having the yes/no values are written into an XML file.
When the UI of the application loads, the fields from the XML files (having yes/no values) are displayed in the symptom part of the application.
(2) Output field generator query
This query is used to retrieve all the fields present in the global schema, that has to be displayed along with the output. The patients’ name, phone number and address serve as default fields in the output as they pave the way to contact the patients and get their treatment done. All the attributes parsed by the DOM parser on the XML schema is displayed in the output fields section.
User Input
The doctor selects the COVID-19 symptoms for which the patients having risk should be identified. After selecting the symptoms, the other output fields that have to be displayed are also given as the input.
Result Integrator
The query fired by the doctor is split into two parts
-
i)
Find the patient IDs that have the value ‘yes’ for all the COVID-19 symptoms selected by the doctor.
-
ii)
For the patient IDs obtained from the step (i) retrieve the selected fields with matching records across three different data sources and display them to the doctor.
Implementation
All the fields present in the global schema are read using a DOM parser and displayed in the front end as shown in Fig. 2. The doctor can select the fields to be retrieved for the patients who are at high risk of the selected COVID-19 symptoms.
Once the doctor clicks on retrieve patients button, the patients having the symptoms are retrieved and the selected output fields are displayed in the front end along with the patient’s name, address and contact number. The doctor can now contact the patient and provide them the required medication.
Results and Discussions
Thus, heterogeneous data integration is accomplished using XML-based ontology creation and mapping. The performance of the system is depicted in the form of graphs below.
Schema Generation and Mapping
The time taken to generate the schema from three different data source increases linearly with time and the Graph is depicted in the figure.
The generation of schema and mapping for about 16,000 records took about 15 s, whereas for the time taken for 7500 records is 6 s as shown in Fig. 3.
Symptom Field Generation
The time taken to generate the symptoms by checking the fields having Boolean data values from three different data source increases linearly with time and the graph is depicted below. The symptoms retrieved are written into an XML file. When the user loads the application, a DOM parser fetches those fields and displays them in the browser as shown in Fig. 4.
The symptom generation process took about 12 s for about 16,000 records. whereas for the time taken for 7500 records is 4 s.
Application Startup Time
The application startup duration measures the time elapsed from the moment the kernel launches a new application process to the instant the application displays its main window. A system is said to have a good performance only when its application startup time is low. Low application startup time is directly linked with high performance.
The time taken to deploy the developed application on a tomcat server instance along with the increasing size of the dataset is shown below as shown in Fig. 5.
The application startup time mostly remains constant for increasing dataset sizes.
Retrieval of Patients
This step includes two parts
-
i)
Symptom checking
-
ii)
Output field retrieval
The graph shows a linear increase in time along with the increasing size of the dataset as shown in Fig. 6.
The time taken to retrieve output from a set of 28,000 records took about 6 s, whereas for the time taken for 14,000 records is 4 s. This is much better when compared to the approach proposed in merging three non-relational databases [21], where the result was obtained in about 12 s for the same number of records.
Conclusion
The proposed method enables data integration from three different data sources i.e., Excel, SQL Server and MongoDB using an XML schema-based ontology and the performance of the system is also good. Moreover, this technique adheres to storage optimization principles. Instead of storing the entire data from the dataset, we extract the schema and handle the queries according to global schema and mapping. Another advantage of this method is that, the model does not suffer from insert, update and delete anomalies, since the mapping is done according to the schema and not data. This method can also be further extended by creating ontologies in RDF/Turtle formats, retrieving data using SPARQL and evaluating its performance.
References
Hammad R, Barhoush M, Abed-Alguni BH. A semantic-based approach for managing healthcare big data: a survey. J Healthc Eng. 2020. https://doi.org/10.1155/2020/8865808.
Li G. Improving biomedical ontology matching using domain-specific word embeddings. In: Proceedings of the 4th International Conference on Computer Science and Application Engineering. Article No. 120; 2020. p. 1–5. https://doi.org/10.1145/3424978.3425102.
Pan H, et al. Biomedical ontologies and their development, management, and applications in and beyond China. J Bio-X Res. 2019;2(4):178–84. https://doi.org/10.1097/jbr.0000000000000051.
Pandey A, Khan A, Abushark Y, Alam M, Agrawal A, Kumar R, Khan R. Key issues in healthcare data integrity: analysis and recommendations. IEEE Access. 2020;8:40612–28.
Zhang Q, Lian B, Cao P, Sang Y, Huang W, Qi L. Multi-source medical data integration and mining for healthcare services. IEEE Access. 2020;8:165010–7.
Asfand-e-yar M, Ali R. Semantic integration of heterogeneous databases of same domain using ontology. IEEE Access. 2020;8:77903–19. https://doi.org/10.1109/ACCESS.2020.2988685.
Li R, Mo T, Yang J, Jiang S, Li T, Liu Y. Ontologies-based domain knowledge modeling and heterogeneous sensor data integration for bridge health monitoring systems. IEEE Trans Ind Inform. 2021;17(1):321–32.
Dong Y, Dragut EC, Meng W. Normalization of duplicate records from multiple sources. IEEE Trans Knowl Data Eng. 2019;31(4):769–82.
Dhayne H, Haque R, Kilany R, Taher Y. In search of big medical data integration solutions—a comprehensive survey. IEEE Access. 2019;7:91265–90.
Peral J, Ferrández A, Gil D, Muñoz-Terol R, Mora H. An ontology-oriented architecture for dealing with heterogeneous data applied to telemedicine systems. In: Special section on ambient intelligence environments with wireless sensor networks from the point of view of big data and smart and sustainable cities, August 15, 2018.
ZhangH, Guo Y, Li Q, George TJ, Shenkman EA, Bian J. Data integration through ontology-based data access to support integrative data analysis: a case study of cancer survival. In: IEEE international conference on bioinformatics and biomedicine (BIBM); 2017.
Zhao S, Qian Q. Ontology based heterogeneous materials database integration and semantic query. AIP Adv. 2017;7(10):105325.
Hazber MAG, Li R, Gu X, Xu G. Integration mapping rules: transforming relational database to semantic web ontology. Appl Math Inf Sci. 2016;10(3):881–901.
Banerjee S, Goto T, Debnath NC, Sarkar A. Ontology driven query language for NoSQL databases. In: IEEE 15th International Conference on Industrial Informatics (INDIN); 2017. p. 951–956, ISBN:978-1-5386-0838-8. https://doi.org/10.1109/INDIN.2017.8104900.
Mahria BB, Chaker I, Zahi A. A novel approach for learning ontology from relational database: from the construction to the evaluation. J Big Data. 2021;8:25. https://doi.org/10.1186/s40537-021-00412-2
Livitckaia K, Koutkias V, Kouidi E, van Gils M, Maglaveras N, Chouvarda I. ‘Optimal’: an ontology for patient adherence modeling in physical activity domain. BMC Med Inform Decis Mak. 2019. https://doi.org/10.1186/s12911-019-0809-9.
Ong E, et al. Modelling kidney disease using ontology: insights from the Kidney Precision Medicine Project. Nat Rev Nephrol. 2020;16(11):686–96. https://doi.org/10.1038/s41581-020-00335-w.
Reyes-Peña C, Tovar M, Bravo M, Motz R. An ontology network for diabetes mellitus in Mexico. J Biomedi Semant. 2021;12(1):19. https://doi.org/10.1186/s13326-021-00252-2.
He Y, et al. CIDO, a community-based ontology for coronavirus disease knowledge and data integration, sharing, and analysis. Sci Data. 2020. https://doi.org/10.1038/s41597-020-0523-6.
Gayathri M, Jagadeesh Kannan R. Ontology based Indian medical system. Mater Today Proc. 2018;5(1):1974–9. https://doi.org/10.1016/j.matpr.2017.11.301.
Kiran VK, Vijayakumar R. Ontology based data integration of NoSQL datastores. In: 9th international conference on industrial and information systems (ICIIS). 2014.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the topical collection “Predictive Artificial Intelligence for Cyber Security and Privacy” guest edited by Hardik A. Gohel, S. Margret Anouncia and Anthoniraj Amalanathan.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Thirumahal, R., Sudha Sadasivam, G. & Shruti, P. Semantic Integration of Heterogeneous Data Sources Using Ontology-Based Domain Knowledge Modeling for Early Detection of COVID-19. SN COMPUT. SCI. 3, 428 (2022). https://doi.org/10.1007/s42979-022-01298-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42979-022-01298-4