INTRODUCTION
Staphylococcus aureus is a leading cause of infections in the USA and is associated with substantial morbidity and mortality [Reference Hidron1–Reference Cosgrove3]. Vancomycin has traditionally been the mainstay of therapy for infections due to methicillin-resistant S. aureus (MRSA) as well as methicillin-susceptible S. aureus (MSSA) in patients unable to tolerate β-lactam antibiotics. However, recent evidence has emerged demonstrating a trend of increasing vancomycin minimum inhibitory concentrations (MICs) in vancomycin-susceptible S. aureus isolates, referred to as vancomycin ‘MIC creep’ [4–Reference Hawser7].
Several studies suggest that infections caused by both MSSA and MRSA isolates with reduced vancomycin susceptibility (RVS), as determined by Etest or broth microdilution methods [8, 9], are associated with higher rates of treatment failure and mortality, particularly in serious infections such as bacteraemia [Reference Hidayat10–Reference Price21]. As such, a clinical prediction rule for RVS would have important implications for the clinical management of S. aureus bacteraemia, both for early recognition of a higher risk of treatment failure in these patients, as well as selection of antimicrobial therapy. To our knowledge, there is only one published study in the literature of a clinical prediction rule for RVS in bacteraemia due to S. aureus [Reference Lubin22], but this was limited to MRSA isolates. Moreover, this study utilized broth microdilution susceptibility testing, which produces vancomycin MIC results that are consistently lower than those reported by Etest [Reference Hsu23–Reference Vaudaux25]. We conducted the present study in order to develop and validate clinical prediction rules for identifying RVS as determined by both Etest and broth microdilution testing in bacteraemia due to MSSA and MRSA.
METHODS
Study design and setting
This retrospective cohort study was conducted at two hospitals in the University of Pennsylvania Health System (UPHS) in Philadelphia: the Hospital of the University of Pennsylvania (HUP), a 725-bed academic tertiary-care medical centre, and Penn Presbyterian Medical Center (PPMC), a 344-bed urban community hospital. The study was approved by the institutional review board of the University of Pennsylvania.
Study population
All inpatients with an episode of S. aureus bacteraemia occurring between 1 December 2007 and 31 May 2009 were identified through the HUP Clinical Microbiology Laboratory, which processes all specimens obtained from patients at HUP and PPMC. For patients with multiple episodes of S. aureus bacteraemia, only the first episode of bacteraemia was included for analysis.
Microbiological identification and susceptibility testing of S. aureus isolates
Identification and susceptibility testing of S. aureus was performed and interpreted according to standard methods [26–Reference Barton28]. Standard susceptibility testing was performed using the Vitek2 instrument method. For the purposes of this study, the vancomycin MIC of the isolates was determined by the Etest using Mueller–Hinton agar (BBL; BD Diagnostic Systems, USA) [8], with RVS defined as a vancomycin MIC >1·0 μg/ml [Reference Kullar19]. In addition, vancomycin MICs were determined for all S. aureus isolates by the broth microdilution method [9], utilizing a susceptibility panel containing half-dilution vancomycin concentrations, custom manufactured by Trek Diagnostic Systems (USA), with RVS defined as a vancomycin MIC ⩾1·0 μg/ml [Reference Moise12].
Data collection
Data were abstracted from the Pennsylvania Integrated Clinical and Administrative Research Database (PICARD), which includes demographic, laboratory, pharmacy, and billing information, and has been used effectively in prior studies of antibiotic use and resistance [Reference Barton28–Reference Lee30]. The following clinical data were collected for all subjects: baseline demographics, origin at the time of hospital admission (i.e. physician referral, transfer from another facility, or admission through the emergency department), healthcare-associated infection (transfer from another institution or date of the first positive culture ⩾48 h from admission), hospital location at the time of infection [i.e. intensive care unit (ICU) or medical floor], prior admission to UPHS in the 30 days prior to the culture date, and the all patient refined-diagnosis related group (APRDRG) risk of mortality and severity of illness scores. The presence of the following comorbid conditions was documented in relation to the date of the positive blood culture: diabetes mellitus, malignancy, renal insufficiency (creatinine ⩾2·0 mg/dl or the requirement of dialysis), solid organ or haematopoietic stem cell transplantation, HIV infection, neutropenia (absolute neutrophil count <500/mm3), and receipt of an immunosuppressive agent, including corticosteroids, in the previous 30 days. Furthermore, chart review was performed to collect data on the presence of complicated infection (i.e. endocarditis, osteomyelitis, septic arthritis, epidural and/or spinal abscess) and the presence of intravascular devices (i.e. intravascular catheter, pacemaker or defibrillator, arteriovenous fistula or graft) prior to the episode of bacteraemia. In addition, the Charlson comorbidity index was calculated for each subject [Reference Quan31].
All inpatient antimicrobial therapy administered during the 30 days prior to the episode of bacteraemia was documented. Antimicrobial therapy was categorized by agent or class, including vancomycin, aminoglycosides, extended-spectrum penicillins (e.g. piperacillin-tazobactam), other penicillins (e.g. penicillin, ampicillin, nafcillin, ampicillin/sulbactam), extended-spectrum cephalosporins (e.g. ceftriaxone, cefotaxime, cefepime), other cephalosporins (e.g. cefazolin), trimethoprim-sulfamethoxazole, fluoroquinolones, macrolides (e.g. azithromycin, erythromycin), tetracyclines (e.g. doxycycline), clindamycin, and metronidazole.
Statistical analysis
Model derivation
Models were derived separately for RVS as defined by (1) Etest and (2) broth microdilution testing. Bivariable analyses were conducted to determine the unadjusted association between RVS and potential predictors. Continuous variables were compared using Student's t test or Wilcoxon rank-sum test and categorical variables were compared using χ2 test or Fisher's exact test. Multiple logistic regression analyses were subsequently performed, with variables with P values <0·20 on bivariable analyses considered for inclusion in the model [Reference Mickey and Greenland32]. Variables with a P value of <0·05 in a backward elimination process were retained in the final model. To double check that no significantly predictive variables were removed during this process, each de-selected variable was tested in turn with the final model and re-introduced into the model if the P value was <0·05. Furthermore, variables with a P value of <0·10 were retained based on the ability of the final model to predict the outcome of interest, as well as the ease of use and availability of the predictor on a clinical basis.
The final regression model was transformed to a simplified integer-based score, with a score for each predictor variable assigned by dividing its β-coefficient by the absolute value of the smallest coefficient in the model and rounding up to the nearest integer. The simplified model was built by including a summation of scores from the presence of all independent predictor variables as a single continuous variable.
Model performance characteristics
The sensitivity, specificity, negative predictive value, and positive predictive value for all possible cut-off values were calculated for the final integer-based score model. The discriminatory ability of the prediction rule in the derivation group was quantified using the C statistic, or the area under the receiver-operating characteristic curve (AUC). Calibration was assessed using the Hosmer–Lemeshow χ2 goodness-of-fit test [Reference Hosmer and Lemeshow33], which evaluates expected and observed probabilities in population deciles. Over-optimism of the prediction characteristics refers to inflated predictive capability due to the model-fitting data typically performing better than data not used for the model-fitting process. In order to adjust for the over-optimism, a bootstrapping procedure for ‘optimism’ adjustment [Reference Harrell, Lee and Mark34–Reference Steyerberg36] was applied, based on 10 000 bootstraps. This procedure was used to calculate the optimism-adjusted C statistic for both the final model with all independent predictors, as well as the simplified model with the total score as the single predictor.
Model validation
The prediction rule was validated internally using the bootstrap method for over-optimism in the original derivation population [Reference Efron and Tibshirani35, Reference Steyerberg36], with calculation of the C statistic as in the derivation process.
A two-tailed P value of <0·05 was considered significant. All statistical calculations were performed using commercially available software (Stata version 11.0; StataCorp LP, USA).
RESULTS
Study population
A total of 392 patients with discrete episodes of S. aureus bacteraemia were identified during the study period. There was a significant, although weak, correlation between vancomycin Etest MICs and vancomycin broth microdilution MICs (Spearman's correlation = 0·40, P < 0·001).
The distribution of vancomycin MICs among isolates as determined by Etest was as follows: 17 (4·3%) with MIC ⩽ 0·5 μg/ml, 83 (21·2%) with MIC = 0·75 μg/ml, 158 (40·3%) with MIC = 1·0 μg/ml, 123 (31·4%) with MIC = 1·5 μg/ml, and 11 (2·8%) with MIC = 2·0 μg/ml [Reference Anthony37]. Accordingly, 34·2% of the S. aureus bloodstream isolates demonstrated RVS as defined by a vancomycin MIC >1·0 μg/ml by Etest. The distribution of vancomycin MICs among isolates as determined by broth microdilution testing was as follows: one (0·3%) with MIC = 0·25 μg/ml, 78 (19·9%) with MIC = 0·5 μg/ml, 240 (61·2%) with MIC = 0·75 μg/ml, 62 (15·8%) with MIC = 1·0 μg/ml, and 11 (2·8%) with MIC = 1·5 μg/ml. Accordingly, 73 (18·6%) had RVS as defined by a broth microdilution MIC ⩾ 1·0 μg/ml.
Derivation of the prediction rules
RVS defined by Etest
Significant univariable predictors of RVS as defined by Etest are shown in Table 1a. Isolates with RVS were significantly more likely to be methicillin resistant (P = 0·02) and associated with bacteraemia in patients who had received antibiotics in the 30 days preceding the culture date (P < 0·001), including an aminoglycoside (P = 0·04) or a fluoroquinolone (P = 0·003). The multivariable prediction model, including regression coefficients, adjusted odds ratios (ORs), and assigned point value for the integer-base score, is shown in Table 2a. The final model incorporated four independent predictors of RVS: methicillin resistance, non-white race, healthcare-associated infection, and receipt of any antimicrobial therapy ⩽30 days prior to the first positive blood culture date. A total score in the simplified model is therefore calculated by summation of individual point values of all predictors that are present for a given patient, with possible scores ranging from −2 to 4.
Table 1. Unadjusted variables associated with RVS as defined by (a) Etest and (b) broth microdilution test in Staphylococcus aureus bacteraemia

RVS, Reduced vancomycin susceptibility; OR, odds ratio; CI, confidence interval; ICU, intensive-care unit; UPHS, University of Pennsylvania Health System; APRDRG, all patient refined-diagnosis related group.
Only those variables with a P value ⩽0·30 are shown.
* All other agents and classes of antimicrobials not shown due to P values >0·20 on univariable analyses.
Table 2. Multivariable prediction model of RVS as defined by (a) Etest and (b) broth microdilution test in Staphylococcus aureus bacteraemia (n = 392)

RVS, Reduced vancomycin susceptibility; OR, odds ratio; CI, confidence interval.
RVS defined by broth microdilution test
Significant univariable predictors of RVS as defined by broth microdilution testing are shown in Table 1b. Isolates with RVS were significantly more likely to be methicillin resistant (P = 0·001) and healthcare-associated (P = 0·04), as well as associated with bacteraemia in patients who were admitted through the emergency department (P = 0·01), had an intravascular device (P = 0·01), and were in the ICU at the time of or ⩽48 h prior to the first positive blood culture date (P = 0·01 and P = 0·07, respectively). The multivariable prediction model, including regression coefficients, adjusted ORs, and assigned point value for the integer-base score, is shown in Table 2b. The final model incorporated four independent predictors of RVS: methicillin resistance, admission through the emergency department, presence of an intravascular device, and malignancy. A total score is calculated in the same fashion as described for RVS as defined by Etest, with possible scores ranging from −2 to 2.
Discrimination, calibration, and validation
RVS defined by Etest
The clinical prediction rule for RVS as determined by Etest with four independent predictors demonstrated good discrimination (C statistic = 0·642, optimism-adjusted C statistic = 0·625) and was well-calibrated (Hosmer–Lemeshow χ2 = 8·97, P = 0·25). When the prediction rule was applied to the validation cohort, the C statistic was 0·650. No collinear relationships were detected in the final model, with variation inflation factors for the variables ranging from 1·03–1·39.
The point-based prediction rule with total score as the single predictor also demonstrated good discrimination (C statistic = 0·646, optimism-adjusted C statistic = 0·645) and good calibration (Hosmer–Lemeshow χ2 = 2·77, P = 0·60). Performance characteristics of the simplified prediction model are shown in Table 3. At the cut-off value of ⩾0, the clinical prediction rule demonstrated a sensitivity of 78·4% and a specificity of 42·6%. Similarly, at the cut-off value of ⩾1, the clinical prediction rule demonstrated a sensitivity of 47·8% and a specificity of 73·3%.
Table 3. Performance characteristics of the simplified prediction model for RVS as defined by Etest in Staphylococcus aureus bacteraemia
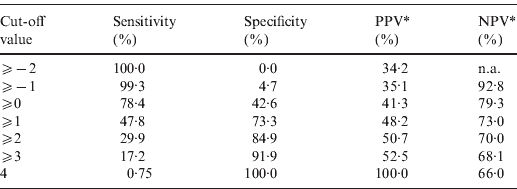
RVS, Reduced vancomycin susceptibility; PPV, positive predictive value; NPV, negative predictive value; n.a., not applicable.
* As calculated given a prevalence of RVS of 34·2%.
RVS defined by broth microdilution test
The clinical prediction rule for RVS as defined by broth microdilution test with four independent predictors demonstrated good discrimination (C statistic = 0·693, optimism-adjusted C statistic = 0·676) and was well-calibrated (Hosmer–Lemeshow χ2 = 10·9, P = 0·15). When the prediction rule was applied to the validation cohort, the C statistic was 0·701. No collinear relationships were detected in the final model, with variation inflation factors for the variables ranging from 1·03 to 1·05.
The point-based prediction rule with total score as the single predictor also demonstrated good discrimination (C statistic = 0·684, optimism-adjusted C statistic = 0·683) and good calibration (Hosmer–Lemeshow χ2 = 2·25, P = 0·32). Performance characteristics of the simplified prediction model are shown in Table 4. At the cut-off value of ⩾0, the clinical prediction rule demonstrated a sensitivity of 89·0% and a specificity of 27·0%. Similarly, at the cut-off value of ⩾1, the clinical prediction rule demonstrated a sensitivity of 61·6% and a specificity of 71·2%.
Table 4. Performance characteristics of the simplified prediction model for RVS as defined by broth microdilution test in Staphylococcus aureus bacteraemia
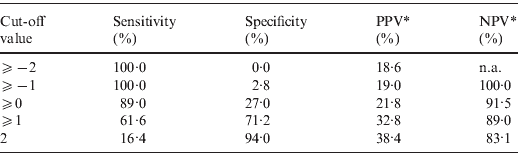
RVS, Reduced vancomycin susceptibility; PPV, positive predictive value; NPV, negative predictive value; n.a., not applicable.
* As calculated given a prevalence of RVS of 18·6%.
DISCUSSION
The results of our study suggest that a simplified, integer-based scoring model that uses readily available data can identify the presence of RVS in the setting of bacteraemia due to S. aureus. Both prediction rules demonstrated good calibration and discrimination, as well as cut-off values with reasonable sensitivity and specificity and excellent negative predictive values for identifying RVS in both MSSA and MRSA isolates causing bacteraemia.
Multiple studies have demonstrated poor clinical outcomes, including increased mortality, in serious infections due to S. aureus with RVS as defined by either Etest or broth microdilution testing [Reference Hidayat10–Reference Price21]. As such, the ability to permit earlier identification of S. aureus isolates with RVS in the context of bacteraemia would have important clinical implications. The majority of clinical laboratories utilize automated systems for routine susceptibility testing to determine vancomycin MICs, and these do not accurately reflect those obtained using Etest or broth microdilution methods [Reference Hsu23]. A simple clinical prediction rule would therefore allow for more rapid identification of RVS using readily available clinical and microbiological information. Early recognition of RVS would allow for improved risk stratification to identify patients with an increased risk of worse clinical outcomes, including greater mortality, in the context of S. aureus bacteraemia. This information would be an important adjunct in the overall clinical decision-making process, including early consideration of alternative therapy in patients with worsening clinical status on vancomycin, as well as more aggressive interventions such as removal of intravascular devices. For example, a recent study demonstrated improved outcomes with daptomycin compared to vancomycin treatment in patients with MRSA bacteraemia characterized by elevated vancomycin MICs [Reference Moore38]. Finally, given how critical the early receipt of appropriate antibiotic therapy is for improving outcomes in serious infections, including in bacteraemia due to S. aureus [Reference Lodise39, Reference Paul40], awareness of a high likelihood of RVS in S. aureus-associated bacteraemia may aid physicians in the selection of initial empirical antimicrobial treatment.
To our knowledge, there is only one study in the literature describing a clinical prediction rule for RVS in S. aureus bacteraemia [Reference Lubin22]. However, this was limited to methicillin-resistant isolates and the use of broth microdilution testing to define RVS. Interestingly, the prediction rule for RVS as defined by Etest in the present study generated a different set of predictors compared to the prediction rule using broth microdilution methods, with the exception of methicillin resistance. Previous studies have demonstrated poor correlation between the Etest and broth microdilution MIC methods, with the Etest providing consistently higher MIC results [Reference Hsu23, Reference Vaudaux25], and this probably contributed to the resulting difference in predictors in our final models. Nevertheless, many of the predictors in both models are indicators of healthcare exposure, including presence of an intravascular device, prior antimicrobial use, healthcare-associated infection, and malignancy. Given that prior studies demonstrating increased mortality with RVS in S. aureus infections have used both Etest and broth microdilution methods to determine RVS, the availability of prediction rules utilizing either of these methods is of significant clinical utility. As such, while there is still uncertainty regarding the optimal approach for testing for RVS, we would recommend that clinicians base their selection of a prediction rule on the method used to determine vancomycin MICs at their institution, as well as defining characteristics of the patient population under treatment and availability of information.
In addition, to our knowledge, the present study is the first to derive and validate clinical prediction rules for RVS in bacteraemia due to MSSA and MRSA. While data on the impact of RVS on clinical outcomes in infections due to MSSA are limited compared to that for MRSA, currently available studies in the literature demonstrate that RVS is associated with increased mortality in bacteraemia due to methicillin-sensitive isolates [Reference Aguado17, Reference Holmes20, Reference Price21]. As such, the ability to identify RVS in both MSSA and MRSA isolates causing bacteraemia will be important, particularly in the context of data suggesting increased microbial fitness of MSSA strains with RVS compared to MRSA strains with RVS [Reference Ender41].
Interestingly, the ORs for some predictors in both models suggested a potentially protective effect on developing S. aureus bacteraemia with an isolate characterized by RVS (e.g. malignancy and non-white race). While it is possible that host and genetic factors may in part explain these findings, further research is needed to elucidate biological mechanisms that may lead to a decreased risk of infection or colonization with both MSSA and MRSA characterized by RVS.
Our clinical prediction models using broth microdilution and Etest methods demonstrated good discrimination and calibration for identifying RVS in the context of bacteraemia due to MSSA and MRSA. Furthermore, there was no significant loss of discriminatory ability in the validation cohort relative to the derivation population, therefore minimizing the possibility of overfitting and loss of reliability in samples other than our specific dataset [Reference Harrell42]. Both integer-based prediction rules are comprised of four dichotomous predictors, thereby facilitating rapid and straightforward calculation of a total score. The prediction rules each have three demographic or clinical variables that are easily determined by clinicians, as well as the laboratory-based variable of methicillin resistance, which is often available early on during the clinical course. Finally, the selection of a particular cut-off value for the two clinical prediction rules will largely be determined by the clinical context. For example, in situations where missing and/or inappropriately treating a specific infection would be particularly detrimental (i.e. a ‘rule out’ situation), it will be important to ensure the use of a clinical prediction rule with a high negative predictive value (i.e. selecting the cut-off values of ⩾ − 1, and ⩾ − 1 or ⩾0 for RVS as determined by Etest and broth microdilution test, respectively).
Finally, there are several potential limitations of the present study. Our clinical prediction rules were derived from a cohort of patients in a single healthcare system, and the external validity of these models needs to be evaluated before being applied to other clinical settings. Furthermore, studies demonstrating poor clinical outcomes in bacteraemia due to S. aureus with RVS have used a range of MIC cut-offs to determine RVS, and the utility of our clinical prediction rules are dependent on the specific Etest and broth microdilution vancomycin MIC values we used to characterize RVS. Finally, as with all similar clinical prediction rules, selection of antimicrobial therapy should ultimately depend on the specific clinical situation and judgement of the treating physicians.
In sum, we have developed and validated simple clinical prediction rules for RVS in the setting of bacteraemia due to MSSA and MRSA, and specifically using both broth microdilution and Etest methods to determine vancomycin MICs. Although host and organism factors leading to worsened clinical outcomes in RVS, as well as optimal treatment strategies for associated infections still need to be further elucidated, having an easy and rapid clinical prediction rule for early identification of RVS can be used to help guide the timely and individualized management of these serious infections.
ACKNOWLEDGEMENTS
This work was supported by the National Institutes of Health (K24 AI080942 to E.L.) and a Commonwealth Universal Research Enhancement Program grant from the Pennsylvania State Department of Health (to E.L.). The dataset on which this study was based was constructed for a prior study supported by a research grant from Cubist Pharmaceuticals (E.L.). The current study was not supported by Cubist Pharmaceuticals. The funding sources had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; or preparation, review, or approval of the manuscript.
DECLARATION OF INTEREST
E.L. has received research grant support from Merck, AstraZeneca and 3M.








