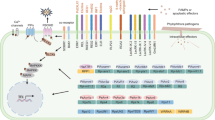
Abstract
Protein-protein interactions have emerged as key determinants of whether plant encounters with pathogens result in disease or successful plant defense. Genetic interactions between plant resistance genes and pathogen avirulence genes enable pathogen recognition by plants and activate plant defense. These gene-for-gene interactions in some cases have been shown to involve direct interactions of the products of the genes, and have indicated plant intracellular localization for certain avirulence proteins. Incomplete specificity of some of the interactions in laboratory assays suggests that additional proteins might be required to confer specificity in the plant. In many cases, resistance and avirulence protein interactions have not been demonstrable, and in some cases, other plant components that interact with avirulence proteins have been found. Investigation to date has relied heavily on biochemical and cytological methods including in vitrobinding assays and immunoprecipitation, as well as genetic tools such as the yeast two-hybrid system. Observations so far, however, point to the likely requirement for multiple, interdependent protein associations in pathogen recognition, for which these techniques can be insufficient. This article reviews the protein-protein interactions that have been described in pathogen recognition by plants, and provides examples of how rapid future progress will hinge on the adoption of new and developing technologies.
Similar content being viewed by others
References
Aronheim, A. 1997. The Sos-recruitment assay: a new type of two-hybrid system. Trends Cell Biol. 7: 21.
Bogdanove, A.J. and Martin, G.B. 2000. AvrPto-dependent Pto-interacting proteins and AvrPto-interacting proteins in tomato. Proc. Natl. Acad. Sci. USA 97: 8836–8840.
Boyes, D.C., Nam, J. and Dangl, J.L. 1998. The Arabidopsis thaliana RPM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hypersensitive response. Proc. Natl. Acad. Sci. USA 95: 15849–15854.
Brent, R. and Finley, R.L. Jr. 1997. Understanding gene and allele function with two-hybrid methods. Annu. Rev. Genet. 31: 663–704.
Bryan, G.T., Wu, K.S., Farrall, L., Jia, Y., Hershey, H.P., McAdams, S.A., Faulk, K.N., Donaldson, G.K., Tarchini, R. and Valent, B. 2000. A single amino acid difference distinguishes resistant and susceptible alleles of the rice blast resistance gene Pi-ta. Plant Cell 12: 2033–2046.
Chang, J.H., Rathjen, J.P., Bernal, A.J., Staskawicz, B.J. and Michelmore, R.W. 2000. avrPto enhances growth and necrosis caused by Pseudomonas syringae pv. tomato in tomato lines lacking either Pto or Prf. Mol. Plant-Microbe Interact. 13: 568–571.
Chen, J.D. and Evans, R.M. 1995. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature 377: 454–457.
Dang, C.V., Barrett, J., Villa-Garcia, M., Resar, L.M., Kato, G.J. and Fearon, E.R. 1991. Intracellular leucine zipper interactions suggest c-Myc hetero-oligomerization. Mol. Cell Biol. 11: 954–962.
Dangl, J.L. and Jones, J.D. 2001. Plant pathogens and integrated defence responses to infection. Nature 411: 826–833.
de Wit, P.J., Lauge, R., Honee, G., Joosten, M.H., Vossen, P., Kooman-Gersmann, M., Vogelsang, R. and Vervoort, J.J. 1997. Molecular and biochemical basis of the interaction between tomato and its fungal pathogen Cladosporium fulvum. Antonie Van Leeuwenhoek 71: 137–141.
Dove, S.L., Joung, J.K. and Hochschild, A. 1997. Activation of prokaryotic transcription through arbitrary protein-protein contacts. Nature 386: 627–630.
Ellis, J.G., Lawrence, G.J., Luck, J.E. and Dodds, P.N. 1999. Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell 11: 495–506.
Feys, B.J., Moisan, L.J., Newman, M.A. and Parker, J.E. 2001. Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J 20: 5400–5411.
Finley, R.L. Jr. and Brent, R. 1994. Interaction mating reveals binary and ternary connections between Drosophila cell cycle regulators. Proc. Natl. Acad. Sci. USA 91: 12980–12984.
Flor, H.H. 1955. Host-parasite interaction in flax rust-its genetics and other implications. Phytopathology 45: 680–685.
Frederick, R.D., Thilmony, R.L., Sessa, G. and Martin, G.B. 1998. Recognition specificity for the bacterial avirulence protein AvrPto is determined by Thr-204 in the activation loop of the tomato Pto kinase. Mol. Cell 2: 241–245.
Galan, J.E. and Collmer, A. 1999. Type III secretion machines: bacterial devices for protein delivery into host cells. Science 284: 1322–1328.
Gopalan, S., Bauer, D.W., Alfano, J.R., Loniello, A.O., He, S.Y. and Collmer, A. 1996. Expression of the Pseudomonas syringae avirulence protein AvrB in plant cells alleviates its dependence on the hypersensitive response and pathogenicity (Hrp) secretion system in eliciting genotype-specific hypersensitive cell death. Plant Cell 8: 1095–1105.
Grant, M. and Mansfield, J. 1999. Early events in host-pathogen interactions. Curr. Opin. Plant Biol. 2: 312–319.
Jia, Y., McAdams, S.A., Bryan, G.T., Hershey, H.P. and Valent, B. 2000. Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19: 4004–4014.
Keen, N.T. 1990. Gene-for-gene complementarity in plant-pathogen interactions. Annu. Rev. Genet. 24: 447–463.
Kim, Y.-J., Lin, N.-C. and Martin, G.B. 2002. Two distinct Pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell 109: in press.
Kobe, B. and Deisenhofer, J. 1995. Proteins with leucine-rich repeats. Curr. Opin. Struct. Biol. 5: 409–416.
Kooman-Gersmann, M., Honee, G., Bonnema, G. and de Wit, P.J.G.M. 1996. A high-affinity binding site for the AVR9 peptide elicitor of Cladosporium fulvum is present on plasma membranes of tomato and other solanaceous plants. Plant Cell 8: 929–938.
Kooman-Gersmann, M., Vogelsang, R., Vossen, P., van den Hooven, H.W., Mahe, E., Honee, G. and de Wit, P.J. 1998. Correlation between binding affinity and necrosis-inducing activity of mutant AVR9 peptide elicitors. Plant Physiol. 117: 609–618.
Leister, R.T. and Katagiri, F. 2000. A resistance gene product of the nucleotide binding site: leucine rich repeats class can form a complex with bacterial avirulence proteins in vivo. Plant J. 22: 345–354.
Leister, R.T., Ausubel, F.M. and Katagiri, F. 1996. Molecular recognition of pathogen attack occurs inside of plant cells in plant disease resistance specified by the Arabidopsis genes RPS2 and RPM1. Proc. Natl. Acad. Sci. USA 93: 15497–15502.
Licitra, E.J. and Liu, J.O. 1996. A three-hybrid system for detecting small ligand-protein receptor interactions. Proc. Natl. Acad. Sci USA 93: 12817–12821.
Luderer, R., Rivas, S., Nurnberger, T., Mattei, B., van den Hooven, H.W., van der Hoorn, R.A., Romeis, T., Wehrfritz, J.M., Blume, B., Nennstiel, D., Zuidema, D., Vervoort, J., De Lorenzo, G., Jones, J.D., de Wit, P.J. and Joosten, M.H. 2001. No evidence for binding between resistance gene product Cf-9 of tomato and avirulence gene product AVR9 of Cladosporium fulvum. Mol. Plant-Microbe Interact. 14: 867–876.
Martin, G.B., Brommonschenkel, S.H., Chunwongse, J., Frary, A., Ganal, M.W., Spivey, R., Wu, T., Earle, E.D. and Tanksley, S.D. 1993. Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science 262: 1432–1436.
Mudgett, M.B. and Staskawicz, B.J. 1999. Characterization of the Pseudomonas syringae pv. tomato AvrRpt2 protein: demonstration of secretion and processing during bacterial pathogenesis. Mol. Microbiol. 32: 927–941.
Neubauer, G., King, A., Rappsilber, J., Calvio, C., Watson, M., Ajuh, P., Sleeman, J., Lamond, A. and Mann, M. 1998. Mass spectrometry and EST-database searching allows characterization of the multi-protein spliceosome complex. Nature Genet. 20: 46–50.
Nimchuk, Z., Marois, E., Kjemtrup, S., Leister, R.T., Katagiri, F. and Dangl, J.L. 2000. Eukaryotic fatty acylation drives plasma membrane targeting and enhances function of several type III effector proteins from Pseudomonas syringae. Cell 101: 353–363.
Oldroyd, G.E.D. and Staskawicz, B.J. 1998. Genetically engineered broad-spectrum disease resistance in tomato. Proc. Natl. Acad. Sci.95: 10300–10305.
Orbach, M.J., Farrall, L., Sweigard, J.A., Chumley, F.G. and Valent, B. 2000. A telomeric avirulence gene determines efficacy for the rice blast resistance gene Pi-ta. Plant Cell 12: 2019–2032.
Pandey, A. and Mann, M. 2000. Proteomics to study genes and genomes. Nature 405: 837–846.
Rathjen, J.P., Chang, J.H., Staskawicz, B.J. and Michelmore, R.W. 1999. Constitutively active Pto induces a Prf-dependent hypersensitive response in the absence of avrPto. EMBO J. 18: 3232–3240.
Remy, I. and Michnick, S.W. 1999. Clonal selection and in vivo quantitation of protein interactions with protein-fragment complementation assays. Proc. Natl. Acad. Sci. USA 96: 5394–5399.
Ren, T., Qu, F. and Morris, T.J. 2000. HRT gene function requires interaction between a NAC protein and viral capsid protein to confer resistance to turnip crinkle virus. Plant Cell 12: 1917–1926.
Reuber, T. and Ausubel, F. 1996. Isolation of Arabidopsis genes that differentiate between resistance responses mediated by the RPS2 and RPM1 disease resistance genes. Plant Cell 8: 241–249.
Riely, B.K. and Martin, G.B. 2001. Ancient origin of pathogen recognition specificity conferred by the tomato disease resistance gene Pto. Proc. Natl. Acad. Sci. USA 98: 2059–2064.
Ritter, C. and Dangl, J. 1996. Interference between two specific pathogen recognition events mediated by distinct plant disease resistance genes. Plant Cell 8: 251–257.
Salmeron, J.M., Oldroyd, G.E.D., Rommens, C.M.T., Scofield, S.R., Kim, H.S., Lavelle, D.T., Dahlbeck, D. and Staskawicz, B.J. 1996. Tomato prf is a member of the leucine-rich repeat class of plant disease resistance genes and lies embedded within the Pto kinase gene cluster. Cell 86: 123–133.
Scofield, S.R., Tobias, C.M., Rathjen, J.P., Chang, J.H., Lavelle, D.T., Michelmore, R.W. and Staskawicz, B.J. 1996. Molecular basis of gene-for-gene specificity in bacterial speck disease of tomato. Science 274: 2063–2065.
Shan, L., He, P., Zhou, J.M. and Tang, X. 2000a. A cluster of mutations disrupt the avirulence but not the virulence function of AvrPto. Mol. Plant-Microbe Interact. 13: 592–598.
Shan, L., Thara, V.K., Martin, G.B., Zhou, J. and Tang, X. 2000b. The Pseudomonas AvrPto protein is differentially recognized by tomato and tobacco and is localized to the plant plasma membrane. Plant Cell 12: 2323–2338.
Song, W.Y., Wang, G.L., Chen, L.L., Kim, H.S., Pi, L.Y., Holsten, T., Gardner, J., Wang, B., Zhai, W.X., Zhu, L.H., Fauquet, C. and Ronald, P. 1995. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270: 1804–1806.
Subramaniam, R., Desveaux, D., Spickler, C., Michnick, S.W. and Brisson, N. 2001. Direct visualization of protein interactions in plant cells. Nature Biotechnol. 19: 769–972.
Szurek, B., Marois, E., Bonas, U. and Van, D.A.G. 2001. Eukaryotic features of the Xanthomonas type III effector AvrBs3: protein domains involved in transcriptional activation and the interaction with nuclear import receptors from pepper. Plant J. 26: 523–534.
Tang, X., Frederick, R.D., Zhou, J., Halterman, D.A., Jia, Y. and Martin, G.B. 1996. Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science 274: 2060–2063.
Van den Ackerveken, G., Marois, E. and Bonas, U. 1996. Recognition of the bacterial avirulence protein AvrBs3 occurs inside the host plant cell. Cell 87: 1307–1316.
Van der Biezen, E.A. and Jones, J.D. 1998. Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem. Sci. 23: 454–546.
Vidal, M., Brachmann, R.K., Fattey, A., Harlow, E. and Boeke, J.D. 1996. Reverse two-hybrid and one-hybrid systems t detect dissociation of protein-protein and DNA-protein interactions. Proc. Natl. Acad. Sci. USA 93: 10315–10320.
Wise, R.P. 2000. Disease resistance: what's brewing in barley genomics. Plant Dis. 84: 1160–1170.
Yang, B., Zhu, W., Johnson, L.B. and White, F.F. 2000. The virulence factor AvrXa7 of Xanthomonas oryzae pv. oryzae is a type III secretion pathway-dependent nuclear-localized double-stranded DNA-binding protein. Proc. Natl. Acad. Sci. USA 97: 9807–9812.
Yang, Y. and Gabriel, D.W. 1995. Xanthomonas avirulence-pathogenicity gene family encodes functional plant nuclear targeting signals. Mol. Plant-Microbe Interact. 8: 627–631.
Zhang, Y., Fan, W., Kinkema, M., Li, X. and Dong, X. 1999. Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc. Natl. Acad. Sci. USA 96: 6523–6528.
Zhou, J., Loh, Y.T., Bressan, R.A. and Martin, G.B. 1995. The tomato gene Pti1 encodes a serine-threonine kinase that is phosphorylated by Pto and is involved in the hypersensitive response. Cell 83: 925–935.
Zhou, J., Tang, X. and Martin, G.B. 1997. The Pto kinase conferring resistance to tomato bacterial speck disease interacts with proteins that bind a cis-element of pathogenesis-related genes. EMBO J. 16: 3207–3218.
Zhou, J.M., Trifa, Y., Silva, H., Pontier, D., Lam, E., Shah, J. and Klessig, D.F. 2000. NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol. Plant-Microbe Interact. 13: 191–202.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Bogdanove, A.J. Protein-protein interactions in pathogen recognition by plants. Plant Mol Biol 50, 981–989 (2002). https://doi.org/10.1023/A:1021263027600
Issue Date:
DOI: https://doi.org/10.1023/A:1021263027600