Abstract
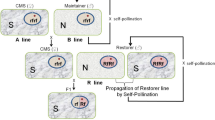
Rice (Oryza sativa L.) productivity is affected by several biotic and abiotic stresses. The genetic variability for some of these stresses is limited in the cultivated rice germplasm. Moreover, changes in insect biotypes and disease races are a continuing threat to increased rice production. There is thus an urgent need to broaden the rice gene pool by introgressing genes for such traits from diverse sources. The wild species of Oryza representing AA, BB, CC, BBCC, CCDD, EE, FF, GG and HHJJ genomes are an important reservoir of useful genes. However, low crossability and limited recombination between chromosomes of cultivated and wild species limit the transfer of such genes. At IRRI, a series of hybrids and monosomic alien addition lines have been produced through embryo rescue following hybridization between rice and several distantly related species. Cytoplasmic male sterility and genes for resistance to grassy stunt virus and bacterial blight have been transferred from A genome wild species into rice. Similarly, genes for resistance to brown planthopper, bacterial blight and blast have also been introgressed across crossability barriers from distanly related species into rice. Some of the introgressed genes have been mapped via linkage to molecular markers. One of the genes Xa-21 introgressed from O. longistaminata has been cloned and physically mapped on chromosome 11 of rice using BAC library and flourescence in-situ hybridization. RFLP analysis revealed introgression from 11 of the 12 chromosomes of C genome species into rice. Introgression has also been obtained from other distant genomes (EE, FF, GG) into rice and in majority of the cases one or two RFLP markers were introgressed. Reciprocal replacement of RFLP alleles of wild species with the alleles of O. sativa indicates alien gene transfer through crossing over. The rapid recovery of recurrent phenotypes in BC2 and BC3 generations from wide crosses is an indication of limited recombination. Further cytogenetic and molecular investigations are required to determine precisely the mechanism of introgression of small chromosome segments from distant genomes in the face of limited homoeologous chromosome pairing. Future research should focus on enhancing recombination between homoeologous chromosomes. Introgression of QTL from wild species should be attempted to increase the yield potential of rice.
Similar content being viewed by others

References
Aggarwal RK, Brar DS, Khush GS: Two new genomes in the Oryza complex identified on the basis of molecular divergence analysis using total genomic DNA hybridization. Mol Gen Genet 254: 1–12 (1996).
Amante-Bordeos A, Sitch LA, Nelson R, Dalmacio RD, Oliva NP, Aswidinnoor H, Leung H: Transfer of bacterial blight and blast resistance from the tetraploid wild rice Oryza minuta to cultivated rice, Oryza sativa.Theor Appl Genet 84: 345–354 (1992).
Brar DS, Dalmacio R, Elloran R, Aggarwal R, Angeles R, Khush GS: Gene transfer and molecular characterization of introgression from wild Oryza species into rice. In: Rice Genetics Ill, pp. 477–486. International Rice Research Institute, Manila, Philippines (1996).
Brar DS, Elloran R, Khush GS: Interspecific hybrids produced through embryo rescue between cultivated and eight wild species of rice. Rice Genet News 8: 91–93 (1991).
Brar DS, Khush GS: Wide hybridization and chromosome manipulation in cereals. In: (Evans DH, Sharp WR, Ammirato PV eds). Handbook of Plant Cell Culture, Vol. 4: Techniques and Applications, pp. 221–263. MacMillan Publ Co., New York, USA (1986).
Brar DS, Khush GS: Wide hybridization for enhancing resistance to biotic and abiotic stresses in rainfed lowland rice. In Fragile Lives in Fragile Ecosystems, pp. 901–910. International Rice Research Institute, Manila, Philippines (1995).
Dalmacio R, Brar DS, Ishii T, Sitch LA, Virmani SS, Khush GS: Identification and transfer of a new cytoplasmic male sterility source from Oryza perennis into indica rice (O. sativa). Euphytica 82: 221–225 (1995).
Dalmacio RD, Brar DS, Virmani SS, Khush GS: Male sterile line in rice (Oryza sativa) developed with O. glumaepatula cytoplasm. IRRN 21(1): 22–23 (1996).
Elloran R, Dalmacio R, Brar DS, Khush GS: Production of backcross progenies from a cross of Oryza sativa× O. granulata.Rice Genet Newsl 9: 39 (1992).
Hu CH: Cytogenetic studies of Oryza officinalis complex. III. The genomic constitution of O. punctata and O. eichingeri.Cytologia 35: 304–318 (1970).
Ikeda R, Khush GS, Tabien RE: A new resistance gene to bacterial blight derived from O. longistaminata.Jpn J Breed 40 (suppl 1): 280–281 (1990).
Ishii T, Brar DS, Multani DS, Khush GS: Molecular tagging of genes for brown planthopper resistance and earliness introgressed from Oryza australiensis into cultivated rice, O. sativa.Genome 37: 217–221 (1994).
Jena KK, Khush GS: Embryo rescue of interspecific hybrids and its scope in rice improvement. Rice Genet Newsl 1: 133–134 (1984).
Jena KK, Khush GS: Introgression of genes from Oryza officinalisWell ex Watt to cultivated rice, O. sativaL. Theor Appl Genet 80: 737–745 (1990).
Jena KK, Khush GS, Kochert G: RFLP analysis of rice (Oryza sativaL.) introgression lines. Theor Appl Genet 84: 608–616 (1992).
Jiang J, Gill BS, Wang GL, Ronald PC, Ward DC: Metaphase and interphase fluorescence in situ hybridization mapping of the rice genome with bacterial artificial chromosomes. Proc Natl Acad Sci USA 92: 4487–4491 (1995).
Katayama T, Onizuka W: Intersectional F1plants from Oryza sativa× O. ridleyi and Oryza sativa× O. meyeriana.Jpn J Genet 54: 43–46 (1979).
Khush GS, Bacalangco E, Ogawa T: A new gene for resistance to bacterial blight from O. longistaminata.Rice Genet Newsl 7: 121–122 (1990).
Khush GS, Brar DS: Overcoming the barriers in hybridization. Theor. Appl. Genet. (Monograph No. 16): 47–61 (1992).
Khush GS, Ling KC, Aquino RC, Aquiero VM: Breeding for resistance to grassy stunt in rice. In Proc. 3rd Intern. Congr. SABRAO, pp. 3–9. Plant Breeding Papers 1[4] Canberra, Australia (1977).
Khush GS, Mackill DJ, Sidhu GS: Breeding rice for resistance to bacterial blight. In Bacterial Blight of Rice, pp. 207–211. International Rice Research Institute, Manila, Philippines (1989).
Li HW, Chen CC, Weng TS, Wuu KD: Cytogenetical studies of Oryza sativaL. and its related species 4. lnterspecific crosses involving O. australiensis with O. sativa and O. minuta.Bot Bull Acad 4: 65–74 (1963).
Lin SC, Yuan LP: A mass screening method for testing grassy stunt disease of rice. Hybrid rice breeding in China, pp. 35–51. In: Innovative approaches to rice improvement. International Rice Research Institute, Manila, Philippines (1980).
Ling KC, Aguiero VM, Lee SH: A mass screening method for testing resistance to grassy stunt disease of rice. Plant Dis Reptr 56: 565–569 (1970).
McCouch SR, Tanksley SD: Development and use of restriction fragment length polymorphism in rice breeding and genetics. In: Khush GS, Toenniessen GH (eds). Rice Biotechnology, pp. 109–133, C.A.B. International Wallingford, U. K. (1991).
Morinaga T: Cytogenetical investigations on Oryza speciesL. In: Rice Genetics and Cytogenetics, pp. 91–102. International Rice Research Institute/Elsevier, Amsterdam (1964).
Multani DS, Jena KK, Brar DS, delos Reyes BC, Angeles ER, Khush GS: Development of monosomic alien addition lines and introgression of genes from Oryza australiensis.Domin. to cultivated rice O. sativaL. Theor Appl Genet 88: 102–109 (1994).
Nayar MM: Origin and cytogenetics of rice. Adv. Genet. 17: 153–292 (1973).
Nezu M, Katayama TC, Kihara H: Genetic study of genus OryzaI. Crossability and chromosomal affinity among 17 species. Seiken Ziho 11: 1–11 (1960).
Ramanujam S: Cytogenetical studies in Oryzeae. III. Cytogenetical behaviour of an interspecific hybrid in Oryza.J. Genet. 35: 223–258 (1937).
Rick CM: Differential zygotic lethality in a tomato species hybrid. Genetics 48: 1498–1507 (1963).
Rick CM: Controlled introgression of chromosomes of Solanum pennelli into Lycopersicon esculentum: segregation and recombination. Genetics 26: 753–768 (1969).
Rick CM: Further studies on segregation and recombination in backcross derivatives of a tomato species hybrid. Biol. Zentralbl 91: 209–220 (1971).
Ronald PC, Albano B, Tabien R, Abenes L, Wu K, McCouch S, Tanksiey SD: Genetic and physical analysis of rice bacterial blight resistance locus, Xa-21.Mol Gen Genet 236: 113–120 (1992).
Ronald PC, Tanksley SD: Genetic and physical mapping of the bacterial blight resistance gene Xa-21.Rice Genet Newsl 8: 142-143 (1991).
Sato S, Sakamoto I, Nakasone S: Location of Ef-1 for earliness on Nishimura's seventh chromosome. Rice Genet Newsl 2: 59–60 (1985).
Shin YB, Katayama T: Cytogenetical studies on the genus Oryza. XI Alien addition lines of O. sativa with a single chromosomes of O. officinalis. Japan J Genet 54: 1–10 (1979).
Sitch LA: Incompatibility barriers operating in crosses of Oryza sativa with related species and genera. In: Gustafson JP (ed).Genetic Manipulation in Plant Improvement II, pp. 77–94. Plenum Press, New York (1990).
Song WY, Wang GL, Chen LL, Kim HS, Pi YL, Holsten T, Gardner J, Wang B, Zhai WX, Zhu LH, Fauquet C, Ronald P: A receptor kinase like protein encoded by the rice disease resistance gene, Xa-21.Science 270: 1804–1806 (1995).
Stephens SG: The cytogenetics of speciation in GossypiumI. Selective elimination of the donor parent genotype in interspecific backcrosses. Genetics 34: 627–637 (1949).
Wang G-L, Holsten TE, Song W-Y, Wang H-P, Ronald PC: Construction of a rice bacterial artificial chromosome library and identification of clones linked to the Xa-21 disease resistance locus. The Plant Journal 7: 525–533 (1995).
Wuu KD, Jui Y, Ly KCL, Chou C, Li HW: Cytogenetical studies of Oryza sativaL. and its related species. 3. Two intersectional hybrids, O. sativaL. × brachyanthaA. Chev et Rochr. Bot Bull Acad Sin 4: 51–59 (1963).
Xiao J, Grandillo S, Ahn SN, McCouch SR, Tanksley SD, Yuan L: Genes from wild rice improve yield. Nature 384: 223–224 (1996).
Yasui H, Iwata N: Production of monosomic alien addition lines of Oryza sativa having a single O. punctata chromosome. Rice Genetics II: 147–155 (1991).
Yuan LP: Advantages and constraints to use of hybrid rice varieties. In: Wilson KJ (ed). Proc lnt Workshop on Apomixis in Rice, pp. 1–4. The Rockefeller Foundation New York and China National Centre for Biotechnology Development, Beijing, China (1993).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Brar, D., Khush, G. Alien introgression in rice. Plant Mol Biol 35, 35–47 (1997). https://doi.org/10.1023/A:1005825519998
Issue Date:
DOI: https://doi.org/10.1023/A:1005825519998