Abstract
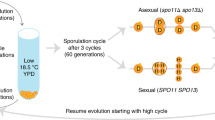
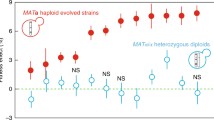
Mutations were accumulated over hundreds of generations in a mutator strain of yeast in a constant laboratory environment. This ensured that mutations were frequent and that the quality of environment remained unchanged. Mutations were accumulated in asexual populations of diploids but their impact on fitness was tested both for the diploid clones and for haploid clones derived from them. Dozens of harmful and lethal mutations accumulated in diploids, but important phenotypic traits, such as maximum growth rate, did not deteriorate by more than 10%. There were no signs of decline in population size. In strong contrast, the populations of haploids derived from the diploids suffered from high mortality; their density was reduced by more than three orders of magnitude. These findings indicate how ineffective natural selection can be in removing deleterious mutations from populations of clonally reproducing diploids. They also suggest that phenotypic assays of heterozygous diploids may be of little value as indicators of increasing genetic degeneration.
Similar content being viewed by others
References
Bell, G., 1982. The Masterpiece of Nature. University of California Press, Berkeley.
Bell, G., 1994. The comparative biology of the alternation of generations. Lect. Math. Life Sci. 25: 1–26.
Boeke, J.D., F. Lacroute & G.R. Fink, 1984. A positive selection for mutants lacking orotidine 5′-phosphate decarboxylase activity in yeast. Mol. Gen. Genet. 197: 345–346.
Charlesworth, B. & K.A. Hughes, 2000. The maintenance of genetic variation in life-history traits, pp. 369–392 in Evolutionary Genetics: From Molecules to Morphology, edited by R.S. Singh & C.B. Krimbas. Cambridge University Press, Cambridge.
Crow, J.F. & M. Kimura, 1970. An introduction to population genetics theory. Harper and Row, New York.
Deng, H.-W. & M. Lynch, 1997. Inbreeding depression and inferred deleterious-mutation parameters in daphnia. Genetics 147: 147–155.
Drake, J.W., B. Charlesworth, D. Charlesworth & J.F. Crow, 1998. Rates of spontaneous mutations. Genetics 148: 1667–1686.
Galitski, T., A.J. Saldanha, C.A. Styles, E.S. Lander & G.R. Fink, 1999. Ploidy regulation of gene expression. Science 285: 251–254.
Garcia-Dorado, A. & A. Caballero, 2000. On the average coefficient of dominance of deleterious spontaneous mutations. Genetics 155: 1991–2001.
Gilligan, D.M., L.M. Woodwarth, M.E. Montgomery et al., 1997. Is mutation accumulation a threat to the survival of endangered populations? Conserv. Biol. 11: 1235–1241.
Haigh, J., 1978. The accumulation of deleterious genes in a population. Theor. Popul. Biol. 14: 251–267.
Haldane, J.B.S., 1924. A mathematical theory of natural and artificial selection: I. Trans. Phil. Soc. 23: 19–41.
Jenkins, C.D. & M. Kirkpatrick, 1995. Deleterious mutation and the evolution of genetic life cycles. Evolution 49: 512–520.
Johnston, M. & D.J. Schoen, 1995. Mutation rates and dominance levels of genes affecting total fitness in two angiosperm species. Science 267: 226–229.
Keightley, P.D. & A. Eyre-Walker, 1999. Terumi Mukai and the riddle of deleterious mutation rates. Genetics 153: 515–523.
Kolodner, R., 1996. Biochemistry and genetics of eukaryotic mismatch repair. Genes Dev. 10: 1433–1442.
Kondrashov, A.S. & J.F. Crow, 1991. Haploidy or diploidy: which is better. Nature 351: 314–315.
Korona, R., 1999a. Genetic load of the yeast Saccharomyces cerevisiaeunder diverse environmental conditions. Evolution 53: 1966–1971.
Korona, R., 1999b. Unpredictable fitness transitions between haploid and diploid strains of genetically loaded yeast Saccharomyces cerevisiae.Genetics 151: 77–85.
Kramer, W., B. Kramer, M.S. Williamson & S. Fogel, 1989. Cloning and nucleotide sequence of DNA mismatch repair gene PMS1 from Saccharomyces cerevisiae: homology of PMS1 to prokaryotic MutL and HexB. J. Bacter. 171: 5339–5346.
Lande, R., 1994. Risk of population extinction from new deleterious mutations. Evolution 48: 1460–1469.
Lynch, M., J. Conery & R. Burger, 1995. Mutational meltdowns in sexual populations. Evolution 49: 1067–1088.
Lynch, M., J. Blanchard, D. Houle, T. Kibota, S. Schultz, L. Vassilieva & J. Villis, 1999. Spontaneous deleterious mutation. Evolution 53: 645–663.
Lynch, M., R. Burger, D. Butcher & W. Gabriel, 1993. The mutational meltdown in asexual populations. Heredity 84: 339–344.
Mable, B. & S.P. Otto}, 1998. The evolution of life cycles with haploid and diploid phases. BioEssays 20: 453–462.
Marsischky, G.T., N. Filosi, M.F. Kane & R. Kolodner, 1996. Redundancy of Saccharomyces cerevisiaeMSH3 and MSH6 in MSH2 dependent mismatch repair. Genes Dev. 10: 407–420.
Mortimer, R.K., 2000. Evolution and variation of the yeast (Saccharomyces) genome. Genome Res. 10: 403–409.
Mukai, T., S.I. Chigusa, L.E. Mettler & J.F. Crow, 1972. Mutation rate and dominance of genes affecting viability in Drosophila melanogaster.Genetics 72: 335–355.
Muller, H.J., 1964. The relation of recombination to mutational advance. Mut. Res. 1: 2–9.
Otto, S.P. & D.B. Goldstein, 1992. Recombination and the evolution of diploidy. Genetics 131: 745–751.
Otto, S.P. & J.C. Marks, 1996. Mating systems and the evolutionary transition between haploidy and diploidy. Biol. J. Linn. Soc. 57: 197–218.
Perrot, V., S. Richerd & M. Valero, 1991. Transition from haploidy to diploidy. Nature 351: 315–317.
Reenan, R.A. & R.D. Kolodner, 1992. Isolation and characterisation of two Saccharomyces cerevisiaegenes encoding homologs of the bacterial HexA and MutS mismatch repair proteins. Genetics 132: 963–973.
Shabalina, S.A., L.Yu. Yampolsky & A.S. Kondrashov, 1997. Rapid decline of fitness in panmictic populations of Drosophila under relaxed selection. Proc. Natl. Acad. Sci. USA 94: 13034–13039.
Sherman, F., 1991. Getting started with yeast, pp. 3–20 in Guide to Yeast Genetics and Molecular Biology, edited by C. Guthrie & G.R. Fink. Academic Press, Inc., London.
Szafraniec, K., R.H. Borts & R. Korona, 2001. Environmental stress and mutational load in diploid strains of the yeast Saccharomyces cerevisiae.Proc. Nat. Acad. Sci. USA 98: 1107–1112.
Szafraniec, K., D.M. Wloch, P. Sliwa, R.H. Borts & R. Korona, 2003. Small fitness effects and weak genetic interactions between deleterious mutations in heterozygous loci of the yeast Saccharomyces cerevisiae.Genet. Res. 82: 19–31.
Vassilieva, L.L., A.M. Hook & M. Lynch, 2000. The fitness effects of spontaneous mutations in Caenorhabditis elegans.Evolution 54: 1234–1246.
Wach, A., A. Brachat, R. Pohlmann & P. Philippsen}, 1994. New heterologous modules for classical and PCR-based disruptions in Saccharomyces cerevisiae}.Yeast 10: 1793–1808.
Williamson, M.S., J. Game & S. Fogel, 1985. Meiotic gene conversion mutants in Saccharomyces cerevisiae.I Isolation and characterization of PMS1-1 and PMS1-2. Genetics 110: 609–646.
Winzeler, E.A., D.D. Shoemaker, A. Astromoff, H. Liang, K. Anderson et al., 1999. Functional characterization of the S. cerevisiaegenome by gene deletion and parallel analysis. Science 285: 901–906.
Wloch, D.M., R.H. Borts & R. Korona, 2001. Epistatic interactions of spontaneous mutations in haploid strains of the yeast Saccharomyces cerevisiae.J. Evol. Biol. 14: 310–316.
Wloch, D.M., K. Szafraniec, R.H. Borts & R. Korona, 2001. Direct estimate of the mutation rate and the distribution of fitness effects in the yeast Saccharomyces cerevisiae.Genetics 159: 441–452.
Zeyl, C., M. Mizesko & A.G.M. de Visser, 2001. Mutational meltdown in laboratory yeast populations. Evolution 55: 909–917.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Sliwa, P., Kluz, J. & Korona, R. Mutational Load and the Transition between Diploidy and Haploidy in Experimental Populations of the Yeast Saccharomyces cerevisiae . Genetica 121, 285–293 (2004). https://doi.org/10.1023/B:GENE.0000039846.12313.98
Issue Date:
DOI: https://doi.org/10.1023/B:GENE.0000039846.12313.98