Abstract
Poly(hydroxyalkanoates) are natural polymers with thermoplastic properties. One polymer of this class with commercial applicability, poly(3-hydroxybutyrate- co-3-hydroxyvalerate) (PHBV) can be produced by bacterial fermentation, but the process is not economically competitive with polymer production from petrochemicals. Poly(hydroxyalkanoate) production in green plants promises much lower costs, but producing copolymer with the appropriate monomer composition is problematic. In this study, we have engineered Arabidopsis and Brassica to produce PHBV in leaves and seeds, respectively, by redirecting the metabolic flow of intermediates from fatty acid and amino acid biosynthesis. We present a pathway for the biosynthesis of PHBV in plant plastids, and also report copolymer production, metabolic intermediate analyses, and pathway dynamics.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Anderson, A.J. & Dawes, E.A. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol. Rev. 54, 450–472 (1990).
Holmes, P.A. Applications of PHB-a microbially produced biodegradable thermoplastic. Phys. Technol. 16, 32–36 (1985).
Poirier, Y., Nawrath, C. & Somerville, C. Production of polyhydroxyalkanoates, a family of biodegradable plastics and elastomers, in bacteria and plants. Biotechnology 13, 142–150 ( 1995).
Steinbüchel, A. Polyhydroxyalkanoic acids. in Biomaterials, novel materials from biological sources (ed. Byrom, D.) 217–230 (Macmillan Publishers Ltd, Basingstoke; 1991).
Byrum, D. Production of poly—hydroxybutyrate:poly—hydroxyvalerate copolymers. FEMS Microbiol. Rev. 102, 247– 250 (1992).
Holmes, P.A., Collins, S.H. & Wright, F. 3-hydroxybutyrate polymers. US Patent no. 4,477,654 (1984).
van der Leij, F.R. & Witholt, B. Strategies for the sustainable production of new biodegradable polyesters in plants: a review. Can. J. Microbiol. 41, 222– 238 (1995).
Nawrath, C., Poirier, Y. & Somerville, C. Targeting of the polyhydroxybutyrate biosynthetic pathway to the plastids of Arabidopsis thaliana results in high levels of polymer accumulation. Proc. Natl. Acad. Sci. USA 91, 12760–12764 (1994).
Poirier, Y., Dennis, D.E., Klomparens, K. & Sommerville, C. Polyhydroxybutyrate, a biodegradable thermoplastic, produced in transgenic plants. Science 256, 520– 523 (1992).
Eschenlauer, A.C., Stoup, S.K., Srienc, F. & Somers, D.A. Production of heteropolymeric polyhydroxyalkanoate in Escherichia coli from a single carbon source. Int. J. Biol. Macromol. 19, 121–130 (1996).
Gruys, K.J. et al. Methods of optimizing substrate pools and biosynthesis of hydroxybutyrate-hydroxyvalerate copolymer in bacteria and plants. PCT Int. Appl., 311 pp. CODEN: PIXXD2. WO 9800557 ( 1998).
Taillon, B.E., Little, R. & Lawther, R.P. Analysis of the functional domains of biosynthetic threonine deaminase by comparison of the amino acid sequences of three wild-type alleles to the amino acid sequence of biodegradative threonine deaminase. Gene 63, 245–252 (1988).
Slater, S. et al. Multiple β-ketothiolases mediate poly(β-hydroxyalkanoate) copolymer synthesis in Ralstonia eutropha. J. Bacteriol. 180, 1979–1987 (1998).
Odell, J.T., Nagy, F. & N.H. Chua. Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature 313, 810–812 (1985).
Broun, P., Boddupalli, S. & Somerville, C. A bifunctional oleate 12-hydroxylase:desaturase from Lesquerella fendleri. Plant J. 13, 201 –210 (1998).
Umbarger, H.E. in Escherichia coli and salmonella: cellular and molecular biology (eds Neidhart, F.C. et al.) 442–457 (ASM Press, Washington, D.C; 1996).
Pledger, W.J. & Umbarger, H.E. Isoleucine and valine metabolism in Escherichia coli. XXI. mutations affecting derepression and valine resistance. J. Bacteriol. 114, 183– 194 (1973).
Burns, R.O., Hofler, J.G. & Luginbuhl, G.H. Threonine deaminase from Salmonella typhimurium . J. Biol. Chem. 254, 1074– 1079 (1979).
Eisenstein, E. et al. An expanded two-state model accounts for homotropic cooperativity in biosynthetic threonine deaminase from Escherichia coli. Biochemistry. 34, 9403–9412 (1995).
Pledger, W.J. & Umbarger, H.E. Isoleucine and valine metabolism in Escherichia coli. XXII. A pleiotropic mutation affecting induction of isomeroreductase activity. J. Bacteriol. 114, 195–207 (1973).
Hagelstein, P., Sieve, B., Klein, M., Jans, H. & Schultz, G. Leucine synthesis in chloroplasts: leucine/isoleucine aminotransferase and valine aminotransferase are different enzymes in spinach chloroplasts. J. Plant Physiol. 150, 23– 30 (1997).
Matthews, B.F. in Plant amino acids: biochemistry and biotechnology. (ed. Singh, B.K.) 205–225 (Marcel Dekker, New York; 1999).
Lydon, J. & Duke, S.O. in Plant amino acids: biochemistry and biotechnology. (ed. Singh, B.K.) 445–464 (Marcel Dekker, New York; 1999).
Lawther, R.P. et al. The complete nucleotide sequence of the ilvGMEDA operon of Escherichia coli K-12. Nucleic Acids Res. 11, 2137–2155 (1987).
Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 ( 1976).
Coruzzi, G., Broglie, R., Edwards, C. & Chua, N.H. Tissue-specific and light-regulated expression of a pea nuclear gene encoding the small subunit of ribulose-1,5-bisphosphate carboxylase. EMBO J. 3 , 1671–1679 (1984).
Bechtold, N., Ellis, J. & Pelletier, G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Ser. III 316, 1194–1199 (1993).
Ye, G., et al. Arabidopsis ovule is the target for Agrobacterium in planta vacuum infiltration transformation. Plant J. 19, 249–257 (1999).
Cashmore, A.R. in Genetic engineering of plants. (eds Kosuge, T., Meredith, C.P. & Hollaender, A.) 29–38 (Plenum Press, New York; 1983).
Fry, J., Barnason, A. & Horsch, R.B. Transformation of Brassica napus with Agrobacterium tumefaciens based vectors. Plant Cell Rep. 6, 321–325 (1987).
Kang, F. & Rawsthorne, S. Starch and fatty acid synthesis in plastids from developing embryos of oilseed rape (Brassica napus L.). Plant J. 6, 795–805 (1994).
Willis, D.E. Automated pre-column derivatization of amino acids with o-phthalaldehyde by a reagent sandwiching technique. J. Chromatogr. 408, 217–225 (1987).
Qureshi, A.A., Elson, C.E. & Lebeck L.A. Application of high-performance liquid chromatography to the determination of glyoxylate synthesis in chick embryo liver. J. Chromatogr. 249, 333–345 (1982).
Koizumi, F., Hideki, A. & Doi, Y. Molecular weight of poly(3-hydroxybutyrate) during biological polymerization in A. eutrophus. Journal of Macromolecular Science, Pure Applied Chemistry. A32, 759–774 (1995).
Acknowledgements
We gratefully acknowledge the excellent technical assistance of Debra Broyles, Laura Casagrande, Kathleen Gonzalez, Catharine Gunter, Brad La Vallee, Susan Lowry, Debbie Mahadeo, Robert Stock, and Gregory Thorne.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
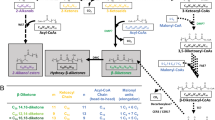
Alternatives for Generation of Propionyl-CoA in planta
Poly(3-hydroxybutyrate-co-3-hydroxyvalerate) (PHBV) can be produced in plants by using the Pyruvate Dehydrogenase Complex (PDC) for converting 2-ketobutyrate to propionyl-CoA. However, under in vivo conditions in transformed plants expressing the ilvA gene, PDC performs this reaction inefficiently, and production of copolymer to high internal concentrations in planta will likely require a supplementary route for conversion of 2-ketobutyrate to propionyl-CoA. There are ways to bypass PDC or supplement its activity, but all require additional transgenes. These routes include modifying the PDC to more readily accept 2-ketobutyrate1, 2, expression of an alternative enzyme system capable of forming propionyl-CoA from 2-ketobutyrate3, or co-expression of a 2-ketoacid oxidase with a propionyl-CoA synthetase or acyl-CoA transferase1, 4.
Propionyl-CoA can also be generated by other routes, although none presents a straightforward alternative to the threonine-derived pathway (See Figure 1). For instance, propionyl-CoA may be generated from acetyl-CoA using a 5-step pathway, part of which is involved in propionyl-CoA degradation in plants5-9. Conversion of acrylyl-CoA to propionyl-CoA is potentially problematic, but an appropriate enzyme may be available from Chroroflexus aurantiacus5. Propionyl-CoA can also be derived from succinyl-CoA using a pathway present in both Rhodococcus ruber and Nocardia corallina10, 11. This pathway is initiated by methylmalonyl-CoA mutase, an enzyme that requires vitamin B12 as a cofactor. However, vitamin B12 is not synthesized in plants9. Rhodococcus and Nocardia also produce minor amounts of 3-hydroxyvaleryl-CoA via a different, uncharacterized route10, 11 that may link to enzymes for degradation of valine and isoleucine. These pathways might also be engineered in plants, but a large number of genes are required.

Figure 1. Potential routes to produce propionyl-CoA in Plants. (GIF 35.5 KB)
Many alternative pathways have the potential to produce propionyl-CoA in plants. However, production of propionyl-CoA from threonine provides the most direct route.
Several other amino acids can be used to produce propionyl-CoA. Methionine, like threonine, generates 2-ketobutyrate during catabolism. This conversion can be performed by a multi-enzyme pathway that also produces cysteine12, or in a single step by L-methionine ?-lyase in a reaction that also produces ammonia and methanethiol13. In either case, supplementation of PDC activity would still be required to efficiently produce propionyl-CoA. Another pathway, present in Clostridium propionicum, converts alanine to propionyl-CoA via lactic acid, lactyl-CoA and acrylyl-CoA14, 15. However, none of the required genes have been cloned, and some of the necessary enzymes are oxygen sensitive16, 17. ß-alanine is another potential starting metabolite for the production of propionyl-CoA18-20. ß-alanine normally plays a critical role as a precursor to Coenzyme-A and acyl carrier protein. However, little is known about the concentration and compartmentalization of ß-alanine in plants, and propionyl-CoA may actually be required for its synthesis.
Literature Cited
-
1
Gruys, K.J., et al. Methods of optimizing substrate pools and biosynthesis of hydroxybutyrate-hydroxyvalerate copolymer in bacteria and plants. PCT Int. Appl., 311 pp. CODEN: PIXXD2. WO 9800557 (1998).
-
2
Inoue, H., et al. Molecular characterization of the mde operon involved in L-methionine catabolism of Pseudomonas putida. J Bacteriol. 179, 3956-3962 (1997).
-
3
Kerscher, L., & Oesterhelt, D. Purification of two 2-oxoacid: ferredoxin oxidoreductases from Halobacterium halobium. Eur. J. Biochem. 116, 587-594 (1981).
-
4
Horswill A.R. & Escalante-Semerena J.C. Propionate catabolism in Salmonella typhimurium LT2: two divergently transcribed units comprise the prp locus at 8.5 centisomes, prpR encodes a member of the sigma-54 family of activators, and the prpBCDE genes constitute an operon. J. Bacteriol. 179, 928-940 (1997).
-
5
Eisenreich, W., Strauss, G., Werz, U., Fuchs, G., & Bacher, A. Retrobiosynthetic analysis of carbon fixation in the phototrophic eubacterium Chloroflexus aurantiacus. Eur. J. Biochem. 215, 619-632 (1993).
-
6
Podkowinski, J., Sroga, G.E., Haselkorn, R., & Gornicki, P. Structure of a gene encoding a cytosolic acetyl-CoA carboxylase of hexaploid wheat. Proc. Natl. Acad. Sci. USA 93, 1870-1874 (1996).
-
7
Preifert, H., & Steinbüchel, A. Identification and molecular characterization of the acetyl coenzyme-A synthetase gene (acoE) of Alcaligenes eutrophus. J. Bacteriol. 174, 6590-6599 (1992).
-
8
Sun, J., Ke, J., Johnson, J.L., Nikolau, B.J., & Wurtele, E.S. Biochemical and molecular biological characterization of CAC2, the Arabidopsis thaliana gene coding for the biotin carboxylase subunit of the plastidic acetyl-coenzyme A carboxylase. Plant Physiol. 115, 1371- 1383 (1997).
-
9
Goodwin, T.W. & Mercer, E.I. Introduction to Plant Biochemistry. 2nd Edition. (Pergamon Press, Oxford, 1985).
-
10
Valentin, H.E., & Dennis, D. Metabolic pathway for poly(3-hydroxybutyrate-co-3-hydroxyvalerate) formation in Nocardia corallina: inactivation of mutB by chromosomal integration of a kanamycin resistance gene. Appl. Environ. Microbiol. 62, 372-379 (1996).
-
11
Williams, D.R., Anderson, A.J., Dawes, E.A. & Ewing, D.F. Production of a co-polyester of 3-hydroxybutyric acid and 3-hydroxyvaleric acid from succinic acid by Rhodococcus ruber: biosynthetic considerations. Appl. Microbiol. Biotechnol. 40, 717-723 (1994).
-
12
Stryer, L. Biochemistry. 3rd Edition. (W. H. Freeman and Co., New York, 1988).
-
13
Tanaka, H., Esaki, N., & Soda, K. A versatile bacterial enzyme: methionine ?-lyase. Enzyme Microb. Technol. 7, 530-537 (1985).
-
14
Schweiger, G., & Buckel, W. On the dehydration of (R)-lactate in the fermentation of alanine to propionate by Clostridium propionicum. FEBS Let. 171, 79-84 (1984).
-
15
Cardon, B.P., & Barker, H.A. Amino acid fermentations by Clostridium propionicum and Diplococcus glycinophilus. Arch. Biochem. Biophys. 12, 165-180 (1947).
-
16
Hofmeister, A.E.M., & Buckel, W. (R)-lactyl-CoA dehydratase from Clostridium propionicum: stereochemistry of the dehydration of (R)-2-hydroxybutyryl-CoA to crotonyl-CoA. Eur. J. Biochem. 206, 547-552 (1992).
-
17
Kuchta, R.D., & Abeles, R.H. Lactate Reduction in Clostridium propionicum: purification and properties of lactyl-CoA dehydratase. J. Biol. Chem. 260, 13181-13189 (1985).
-
18
Arst, H.N., Jr. GABA transaminase provides an alternative route of ß-alanine synthesis in Aspergillus nidulans. Mol. Gen. Genet. 163, 23-27 (1978).
-
19
Kupiecki, R.P., & Coon, M.J. The enzymatic synthesis of ß-aminoisobutyrate, a product of valine metabolism, and of ß-alanine, a product of ß-hydroxypropionate metabolism. J. Biol. Chem. 229, 743-754 (1957).
-
20
Roberts, E., & Bregoff, H.M. Transamination of ?-aminobutyric acid and ß-alanine in brain and liver. J. Biol. Chem. 201, 393-398 (1953).
Rights and permissions
About this article
Cite this article
Slater, S., Mitsky, T., Houmiel, K. et al. Metabolic engineering of Arabidopsis and Brassica for poly(3-hydroxybutyrate- co-3-hydroxyvalerate) copolymer production. Nat Biotechnol 17, 1011–1016 (1999). https://doi.org/10.1038/13711
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/13711
This article is cited by
-
Transgenic plants as a source of polyhydroxyalkanoates
Acta Physiologiae Plantarum (2018)
-
Development and analysis of a highly flexible multi-gene expression system for metabolic engineering in Arabidopsis seeds and other plant tissues
Plant Molecular Biology (2015)