Abstract
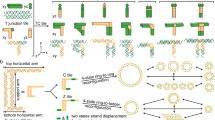
Controlled mechanical movement in molecular-scale devices has been realized in a variety of systems—catenanes and rotaxanes1,2,3, chiroptical molecular switches4, molecular ratchets5 and DNA6—by exploiting conformational changes triggered by changes in redox potential or temperature, reversible binding of small molecules or ions, or irradiation. The incorporation of such devices into arrays7,8 could in principle lead to complex structural states suitable for nanorobotic applications, provided that individual devices can be addressed separately. But because the triggers commonly used tend to act equally on all the devices that are present, they will need to be localized very tightly. This could be readily achieved with devices that are controlled individually by separate and device-specific reagents. A trigger mechanism that allows such specific control is the reversible binding of DNA strands, thereby ‘fuelling’ conformational changes in a DNA machine9. Here we improve upon the initial prototype system that uses this mechanism but generates by-products9, by demonstrating a robust sequence-dependent rotary DNA device operating in a four-step cycle. We show that DNA strands control and fuel our device cycle by inducing the interconversion between two robust topological motifs, paranemic crossover (PX) DNA10,11 and its topoisomer JX2 DNA, in which one strand end is rotated relative to the other by 180°. We expect that a wide range of analogous yet distinct rotary devices can be created by changing the control strands and the device sequences to which they bind.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Pease, A. R. et al. Switching devices based on interlocked molecules. Acc. Chem. Res. 34, 433–444 (2001).
Jimenez, M. C., Dietrich-Buchecker, C. & Sauvage, J.-P. Towards synthetic molecular muscles: contraction and stretching of a linear-rotaxane dimer. Angew. Chem. Int. Edn Engl. 39, 3284–3287 (2000).
Brouwer, A. M. et al. Photoinduction of fast, reversible translational motion in a hydrogen-bonded molecular shuttle. Science 291, 2124–2128 (2001).
Koumura, N., Zijlstra, R. W. J., van Delden, R. A., Harada, N. & Feringa, B. L. Light-driven monodirectional molecular rotor. Nature 401, 152–155 (1999).
Kelly, T. R., De Silva, H. & Silva, R. A. Unidirectional rotary motion in a molecular system. Nature 401, 150–152 (1999).
Mao, C., Sun, W., Shen, Z. & Seeman, N. C. A DNA nanomechanical device based on the B-Z transition. Nature 397, 144–146 (1999).
Winfree, E., Liu, F., Wenzler, L. A. & Seeman, N. C. Design and self-assembly of two-dimensional DNA crystals. Nature 394, 539–544 (1998).
LaBean, T. H. et al. The construction, analysis, ligation and self-assembly of DNA triple crossover complexes. J. Am. Chem. Soc. 122, 1848–1860 (2000).
Yurke, B., Turberfield, A. J., Mills, A. P. Jr, Simmel, F. C. & Neumann, J. L. A DNA-fuelled molecular machine made of DNA. Nature 406, 605–608 (2000).
Seeman, N. C. DNA nicks and nodes and nanotechnology. Nano Lett. 1, 22–26 (2001).
Shen, Z. DNA Polycrossover Molecules and their Applications in Homology Recognition. Thesis, New York Univ. (1999).
Fu, T.-J. & Seeman, N. C. DNA double crossover structures. Biochemistry 32, 3211–3220 (1993).
Seeman, N. C. De novo design of sequences for nucleic acid structure engineering. J. Biomol. Struct. Dyn. 8, 573–581 (1990).
Caruthers, M. H. Gene synthesis machines: DNA chemistry and its uses. Science 230, 281–285 (1985).
Sun, W., Mao, C., Iwasaki, H., Kemper, B. & Seeman, N. C. No braiding of Holliday junctions in positively supercoiled DNA molecules. J. Mol. Biol. 294, 683–699 (1999).
Yang, X., Wenzler, L. A., Qi, J., Li, X. & Seeman, N. C. Ligation of DNA triangles containing double crossover molecules. J. Am. Chem. Soc. 120, 9779–9786 (1998).
Acknowledgements
We thank R. Sha for discussions. This work was supported by the Office of Naval Research, the National Institute of General Medical Sciences, the National Science Foundation/DARPA, the Information Directorate of the Air Force Research Laboratory located at Rome, New York, and the National Science Foundation.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Rights and permissions
About this article
Cite this article
Yan, H., Zhang, X., Shen, Z. et al. A robust DNA mechanical device controlled by hybridization topology. Nature 415, 62–65 (2002). https://doi.org/10.1038/415062a
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/415062a
This article is cited by
-
The biological applications of DNA nanomaterials: current challenges and future directions
Signal Transduction and Targeted Therapy (2021)
-
Self-assembled Möbius strips with controlled helicity
Nature Communications (2020)
-
Framework Nucleic Acids for Cell Imaging and Therapy
Chemical Research in Chinese Universities (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.