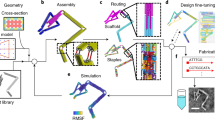
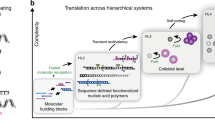
Abstract
DNA is not just the stuff of our genetic code; it is also a means to design self-assembling materials. Grafting DNA onto nano- and microparticles can, in principle, ‘program’ them with information that tells them exactly how to self-assemble. Although fully programmable assembly has not yet been realized, the groundwork has been laid: with an understanding of how specific interparticle attractions arise from DNA hybridization, we can now make systems that reliably assemble in and out of equilibrium. We discuss these advances, and the design rules that will allow us to control — and ultimately program — the assembly of new materials.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Whitesides, G. M., Mathias, J. P. & Seto, C. T. Molecular self-assembly and nanochemistry: a chemical strategy for the synthesis of nanostructures. Science 254, 1312–1319 (1991).
Zhang, S. Fabrication of novel biomaterials through molecular self-assembly. Nat. Biotechnol. 21, 1171–1178 (2003).
Hernandez-Garcia, A. et al. Design and self-assembly of simple coat proteins for artificial viruses. Nat. Nanotechnol. 9, 698–702 (2014).
Bita, I. et al. Graphoepitaxy of self-assembled block copolymers on two-dimensional periodic patterned templates. Science 321, 939–943 (2008).
Ruiz, R. et al. Density multiplication and improved lithography by directed block copolymer assembly. Science 321, 936–939 (2008).
Mai, Y. & Eisenberg, A. Self-assembly of block copolymers. Chem. Soc. Rev. 41, 5969–5985 (2012).
Hu, H., Gopinadhan, M. & Osuji, C. O. Directed self-assembly of block copolymers: a tutorial review of strategies for enabling nanotechnology with soft matter. Soft Matter 10, 3867–3889 (2014).
Glotzer, S. C. & Solomon, M. J. Anisotropy of building blocks and their assembly into complex structures. Nat. Mater. 6, 557–562 (2007).
Sacanna, S., Pine, D. J. & Yi, G.-R. Engineering shape: the novel geometries of colloidal self-assembly. Soft Matter 9, 8096–8106 (2013).
Watson, J. D. & Crick, F. H. C. Molecular structure of nucleic acids: a structure for deoxyribose nucleic acid. Nature 171, 737–738 (1953).
Seeman, N. C. DNA nanotechnology: novel DNA constructions. Annu. Rev. Biophys. Biomol. Struct. 27, 225–248 (1998).
Seeman, N. C. Nucleic acid junctions and lattices. J. Theor. Biol. 99, 237–247 (1982). The first paper to discuss using DNA oligonucleotides as building blocks in 3D self-assembly.
Winfree, E., Liu, F., Wenzler, L. A. & Seeman, N. C. Design and self-assembly of two-dimensional DNA crystals. Nature 394, 539–544 (1998).
Liu, D., Park, S. H., Reif, J. H. & LaBean, T. H. DNA nanotubes self-assembled from triple-crossover tiles as templates for conductive nanowires. Proc. Natl Acad. Sci. USA 101, 717–722 (2004).
Rothemund, P. W. K. et al. Design and characterization of programmable DNA nanotubes. J. Am. Chem. Soc. 126, 16344–16352 (2004).
Zheng, J. et al. From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal. Nature 461, 74–77 (2009).
Ke, Y., Ong, L. L., Shih, W. M. & Yin, P. Three-dimensional structures self-assembled from DNA bricks. Science 338, 1177–1183 (2012). Experimental study showing that multiple copies of prescribed 3D structures can be self-assembled from a ‘soup’ of DNA building blocks with maximally specific interactions; much current research is aimed at duplicating this result with nano- and microparticles.
Ke, Y. et al. DNA brick crystals with prescribed depths. Nat. Chem. 6, 994–1002 (2014).
Rothemund, P. W. K. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006).
Andersen, E. S. et al. Self-assembly of a nanoscale DNA box with a controllable lid. Nature 459, 73–76 (2009).
Douglas, S. M. et al. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 459, 414–418 (2009).
Han, D. et al. DNA origami with complex curvatures in three-dimensional space. Science 332, 342–346 (2011).
Han, D. et al. DNA gridiron nanostructures based on four-arm junctions. Science 339, 1412–1415 (2013).
Zhang, D. Y., Turberfield, A. J., Yurke, B. & Winfree, E. Engineering entropy-driven reactions and networks catalyzed by DNA. Science 318, 1121–1125 (2007).
Zhang, D. Y. & Seelig, G. Dynamic DNA nanotechnology using strand-displacement reactions. Nat. Chem. 3, 103–113 (2011).
Pinheiro, A. V., Han, D., Shih, W. M. & Yan, H. Challenges and opportunities for structural DNA nanotechnology. Nat. Nanotechnol. 6, 763–772 (2011).
Mirkin, C. A., Letsinger, R. L., Mucic, R. C. & Storhoff, J. J. A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature 382, 607–609 (1996).
Alivisatos, A. P. et al. Organization of ‘nanocrystal molecules’ using DNA. Nature 382, 609–611 (1996). References 27 and 28, published concurrently, launched the field of DNA-grafted colloidal particles: reference 27 describes the first reversible DNA-mediated self-assembly of nanoparticles; reference 28 describes the first attempts to program self-assembly of finite structures using DNA.
Jones, M. R., Seeman, N. C. & Mirkin, C. A. Programmable materials and the nature of the DNA bond. Science 347, 1260901 (2015).
Winfree, E. Algorithmic self-assembly of DNA: theoretical motivations and 2D assembly experiments. J. Biomol. Struct. Dyn. 17, 263–270 (2000).
Seeman, N. C. & Lukeman, P. S. Nucleic acid nanostructures: bottom-up control of geometry on the nanoscale. Rep. Prog. Phys. 68, 237–270 (2005).
Geerts, N. & Eiser, E. DNA-functionalized colloids: physical properties and applications. Soft Matter 6, 4647–4660 (2010).
Zhang, X., Wang, R. & Xue, G. Programming macro-materials from DNA-directed self-assembly. Soft Matter 11, 1862–1870 (2015).
Theodorakis, P. E., Fytas, N. G., Kahl, G. & Dellago, C. Self-assembly of DNA-functionalized colloids. Condens. Matter Phys. 18, 22801 (2015).
Jones, M. R. & Mirkin, C. A. Materials science: self-assembly gets new direction. Nature 491, 42–43 (2012).
Hurst, S. J., Lytton-Jean, A. K. R. & Mirkin, C. A. Maximizing DNA loading on a range of gold nanoparticle sizes. Anal. Chem. 78, 8313–8318 (2006).
Valignat, M.-P., Theodoly, O., Crocker, J. C., Russel, W. B. & Chaikin, P. M. Reversible self-assembly and directed assembly of DNA-linked micrometer-sized colloids. Proc. Natl Acad. Sci. USA 102, 4225–4229 (2005).
Dreyfus, R. et al. Simple quantitative model for the reversible association of DNA coated colloids. Phys. Rev. Lett. 102, 048301 (2009).
Kim, A. J., Manoharan, V. N. & Crocker, J. C. Swelling-based method for preparing stable, functionalized polymer colloids. J. Am. Chem. Soc. 127, 1592–1593 (2005).
Wang, Y. et al. Crystallization of DNA-coated colloids. Nat. Commun. 6, 7253 (2015).
Wang, Y. et al. Synthetic strategies toward DNA-coated colloids that crystallize. J. Am. Chem. Soc. 137, 10760–10766 (2015).
Nykypanchuk, D., Maye, M. M., van der Lelie, D. & Gang, O. DNA-guided crystallization of colloidal nanoparticles. Nature 451, 549–552 (2008). Along with reference 72, which was published concurrently, this paper describes the first successful crystallization of DNA-grafted nanoparticles, demonstrating that equilibrium assembly of nanoparticles can be achieved with high DNA surface densities, appropriate sequence design and annealing.
Biancaniello, P., Kim, A. & Crocker, J. Colloidal interactions and self-assembly using DNA hybridization. Phys. Rev. Lett. 94, 058302 (2005). The first experimental study demonstrating that DNA-grafted particles (in this case, microparticles) can crystallize; also contains the first direct measurement of the interaction between DNA-grafted particles and the first coarse-grained theoretical model to quantitatively describe the interaction as a function of temperature.
Xiong, H., van der Lelie, D. & Gang, O. DNA linker-mediated crystallization of nanocolloids. J. Am. Chem. Soc. 130, 2442–2443 (2008).
Yakovchuk, P., Protozanova, E. & Frank-Kamenetskii, M. D. Base-stacking and base-pairing contributions into thermal stability of the DNA double helix. Nucleic Acids Res. 34, 564–574 (2006).
SantaLucia, J. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl Acad. Sci. USA 95, 1460–1465 (1998).
SantaLucia, J. & Hicks, D. The thermodynamics of DNA structural motifs. Annu. Rev. Biophys. Biomol. Struct. 33, 415–440 (2004).
Dirks, R. M. & Pierce, N. A. A partition function algorithm for nucleic acid secondary structure including pseudoknots. J. Comput. Chem. 24, 1664–1677 (2003).
Dimitrov, R. A. & Zuker, M. Prediction of hybridization and melting for double-stranded nucleic acids. Biophys. J. 87, 215–226 (2004).
Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415 (2003).
Markham, N. R. & Zuker, M. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 33, W577–W581 (2005).
Zadeh, J. N. et al. NUPACK: analysis and design of nucleic acid systems. J. Comput. Chem. 32, 170–173 (2011).
Rogers, W. B. & Crocker, J. C. Direct measurements of DNA-mediated colloidal interactions and their quantitative modeling. Proc. Natl Acad. Sci. USA 108, 15687–15692 (2011).
Licata, N. A. & Tkachenko, A. V. Statistical mechanics of DNA-mediated colloidal aggregation. Phys. Rev. E 74, 041408 (2006).
Dreyfus, R. et al. Aggregation–disaggregation transition of DNA-coated colloids: experiments and theory. Phys. Rev. E 81, 041404 (2010).
Varilly, P., Angioletti-Uberti, S., Mognetti, B. M. & Frenkel, D. A general theory of DNA-mediated and other valence-limited colloidal interactions. J. Chem. Phys. 137, 094108 (2012).
Mladek, B. M., Fornleitner, J., Martinez-Veracoechea, F. J., Dawid, A. & Frenkel, D. Quantitative prediction of the phase diagram of DNA-functionalized nanosized colloids. Phys. Rev. Lett. 108, 268301 (2012).
Angioletti-Uberti, S., Varilly, P., Mognetti, B. M., Tkachenko, A. V. & Frenkel, D. Communication: a simple analytical formula for the free energy of ligand–receptor-mediated interactions. J. Chem. Phys. 138, 021102 (2013).
Angioletti-Uberti, S., Varilly, P., Mognetti, B. M. & Frenkel, D. Mobile linkers on DNA-coated colloids: valency without patches. Phys. Rev. Lett. 113, 128303 (2014).
Mognetti, B. M. et al. Predicting DNA-mediated colloidal pair interactions. Proc. Natl Acad. Sci. USA 109, E378–E379 (2012).
Rogers, W. B. & Crocker, J. C. Reply to Mognetti et al.: DNA handshaking interaction data are well described by mean-field and molecular models. Proc. Natl Acad. Sci. USA 109, E380–E380 (2012).
Feng, L., Pontani, L.-L., Dreyfus, R., Chaikin, P. & Brujic, J. Specificity, flexibility and valence of DNA bonds guide emulsion architecture. Soft Matter 9, 9816–9823 (2013).
van der Meulen, S. A. J. & Leunissen, M. E. Solid colloids with surface-mobile DNA linkers. J. Am. Chem. Soc. 135, 15129–15134 (2013).
Tkachenko, A. V. Morphological diversity of DNA-colloidal self-assembly. Phys. Rev. Lett. 89, 148303 (2002).
Xiong, H., van der Lelie, D. & Gang, O. Phase behavior of nanoparticles assembled by DNA linkers. Phys. Rev. Lett. 102, 015504 (2009).
Scarlett, R. T., Crocker, J. C. & Sinno, T. Computational analysis of binary segregation during colloidal crystallization with DNA-mediated interactions. J. Chem. Phys. 132, 234705 (2010).
Scarlett, R. T., Ung, M. T., Crocker, J. C. & Sinno, T. A mechanistic view of binary colloidal superlattice formation using DNA-directed interactions. Soft Matter 7, 1912–1925 (2011).
Martinez-Veracoechea, F. J., Mladek, B. M., Tkachenko, A. V. & Frenkel, D. Design rule for colloidal crystals of DNA-functionalized particles. Phys. Rev. Lett. 107, 045902 (2011).
Vial, S., Nykypanchuk, D., Yager, K. G., Tkachenko, A. V. & Gang, O. Linear mesostructures in DNA–nanorod self-assembly. ACS Nano 7, 5437–5445 (2013).
Li, T. I. N. G., Sknepnek, R., Macfarlane, R. J., Mirkin, C. A. & Olvera de la Cruz, M. Modeling the crystallization of spherical nucleic acid nanoparticle conjugates with molecular dynamics simulations. Nano Lett. 12, 2509–2514 (2012).
Li, T. I. N. G., Sknepnek, R. & Olvera de la Cruz, M. Thermally active hybridization drives the crystallization of DNA-functionalized nanoparticles. J. Am. Chem. Soc. 135, 8535–8541 (2013).
Park, S. Y. et al. DNA-programmable nanoparticle crystallization. Nature 451, 553–556 (2008). Along with reference 42, which was published concurrently, this paper describes the first successful crystallization of DNA-grafted nanoparticles.
Auyeung, E. et al. DNA-mediated nanoparticle crystallization into Wulff polyhedra. Nature 505, 73–77 (2014).
Rogers, W. B., Sinno, T. & Crocker, J. C. Kinetics and non-exponential binding of DNA-coated colloids. Soft Matter 9, 6412–6417 (2013).
Macfarlane, R. J. et al. Nanoparticle superlattice engineering with DNA. Science 334, 204–208 (2011).
Macfarlane, R. J., O’Brien, M. N., Petrosko, S. H. & Mirkin, C. A. Nucleic acid-modified nanostructures as programmable atom equivalents: forging a new “table of elements”. Angew. Chem. Int. Ed. Engl. 52, 5688–5698 (2013).
Casey, M. T. et al. Driving diffusionless transformations in colloidal crystals using DNA handshaking. Nat. Commun. 3, 1209 (2012).
Thaner, R. V. et al. Entropy-driven crystallization behavior in DNA-mediated nanoparticle assembly. Nano Lett. 15, 5545–5551 (2015).
Haxton, T. K., Hedges, L. O. & Whitelam, S. Crystallization and arrest mechanisms of model colloids. Soft Matter 11, 9307–9320 (2015).
Grzelczak, M., Pérez-Juste, J., Mulvaney, P. & Liz-Marzán, L. M. Shape control in gold nanoparticle synthesis. Chem. Soc. Rev. 37, 1783–1791 (2008).
Li, C. et al. High-yield synthesis of single-crystalline gold nano-octahedra. Angew. Chem. Int. Ed. Engl. 119, 3328–3332 (2007).
Kim, D. Y. et al. Seed-mediated synthesis of gold octahedra in high purity and with well-controlled sizes and optical properties. Chem. Eur. J. 17, 4759–4764 (2011).
Hong, S., Shuford, K. L. & Park, S. Shape transformation of gold nanoplates and their surface plasmon characterization: triangular to hexagonal nanoplates. Chem. Mater. 23, 2011–2013 (2011).
Lohse, S. E. & Murphy, C. J. The quest for shape control: a history of gold nanorod synthesis. Chem. Mater. 25, 1250–1261 (2013).
Kraft, D. J. et al. Surface roughness directed self-assembly of patchy particles into colloidal micelles. Proc. Natl Acad. Sci. USA 109, 10787–10792 (2012).
Manoharan, V. N., Elsesser, M. T. & Pine, D. J. Dense packing and symmetry in small clusters of microspheres. Science 301, 483–487 (2003).
Duguet, E., Désert, A., Perro, A. & Ravaine, S. Design and elaboration of colloidal molecules: an overview. Chem. Soc. Rev. 40, 941–960 (2011).
Sindoro, M., Yanai, N., Jee, A.-Y. & Granick, S. Colloidal-sized metal–organic frameworks: synthesis and applications. Acc. Chem. Res. 47, 459–469 (2014).
Rossi, L. et al. Cubic crystals from cubic colloids. Soft Matter 7, 4139–4142 (2011).
Sacanna, S., Irvine, W. T. M., Chaikin, P. M. & Pine, D. J. Lock and key colloids. Nature 464, 575–578 (2010).
Sacanna, S. et al. Shaping colloids for self-assembly. Nat. Commun. 4, 1688 (2013).
Fan, J. A. et al. Self-assembled plasmonic nanoparticle clusters. Science 328, 1135–1138 (2010).
Fan, J. A. et al. DNA-enabled self-assembly of plasmonic nanoclusters. Nano Lett. 11, 4859–4864 (2011).
Kuzyk, A. et al. DNA-based self-assembly of chiral plasmonic nanostructures with tailored optical response. Nature 483, 311–314 (2012).
Kuzyk, A. et al. Reconfigurable 3D plasmonic metamolecules. Nat. Mater. 13, 862–866 (2014).
Sun, D. et al. Light-harvesting nanoparticle core–shell clusters with controllable optical output. ACS Nano 9, 5657–5665 (2015).
Kruglova, O., Demeyer, P.-J., Zhong, K., Zhou, Y. & Clays, K. Wonders of colloidal assembly. Soft Matter 9, 9072–9087 (2013).
Hallam, B. T., Hiorns, A. G. & Vukusic, P. Developing optical efficiency through optimized coating structure: biomimetic inspiration from white beetles. Appl. Opt. 48, 3243–3249 (2009).
Wiersma, D. S. Disordered photonics. Nat. Photonics 7, 188–196 (2013).
Anderson, V. J. & Lekkerkerker, H. N. W. Insights into phase transition kinetics from colloid science. Nature 416, 811–815 (2002).
Soto, C. M. Srinivasan, A. & Ratna, B. R. Controlled assembly of mesoscale structures using DNA as molecular bridges. J. Am. Chem. Soc. 124, 8508–8509 (2002).
Schade, N. B. et al. Tetrahedral colloidal clusters from random parking of bidisperse spheres. Phys. Rev. Lett. 110, 148303 (2013).
Urzhumov, Y. A. et al. Plasmonic nanoclusters: a path towards negative-index metafluids. Opt. Express 15, 14129–14145 (2007).
Alu, A. & Engheta, N. The quest for magnetic plasmons at optical frequencies. Opt. Express 17, 5723–5730 (2009).
Di Michele, L. et al. Multistep kinetic self-assembly of DNA-coated colloids. Nat. Commun. 4, 2007 (2013).
Kim, A. J., Scarlett, R., Biancaniello, P. L., Sinno, T. & Crocker, J. C. Probing interfacial equilibration in microsphere crystals formed by DNA-directed assembly. Nat. Mater. 8, 52–55 (2009).
Jenkins, I. C., Casey, M. T., McGinley, J. T., Crocker, J. C. & Sinno, T. Hydrodynamics selects the pathway for displacive transformations in DNA-linked colloidal crystallites. Proc. Natl Acad. Sci. USA 111, 4803–4808 (2014).
Demirörs, A. F. et al. Long-ranged oppositely charged interactions for designing new types of colloidal clusters. Phys. Rev. X 5, 021012 (2015).
Shevchenko, E. V., Talapin, D. V., Kotov, N. A., O’Brien, S. & Murray, C. B. Structural diversity in binary nanoparticle superlattices. Nature 439, 55–59 (2006).
Mao, C., LaBean, T. H., Reif, J. H. & Seeman, N. C. Logical computation using algorithmic self-assembly of DNA triple-crossover molecules. Nature 407, 493–496 (2000).
Rothemund, P. W. K., Papadakis, N. & Winfree, E. Algorithmic self-assembly of DNA Sierpinski triangles. PLoS Biol. 2, e424 (2004).
Seelig, G., Soloveichik, D., Zhang, D. Y. & Winfree, E. Enzyme-free nucleic acid logic circuits. Science 314, 1585–1588 (2006).
Qian, L. & Winfree, E. Scaling up digital circuit computation with DNA strand displacement cascades. Science 332, 1196–1201 (2011).
Jacobs, W. M., Reinhardt, A. & Frenkel, D. Rational design of self-assembly pathways for complex multicomponent structures. Proc. Natl Acad. Sci. USA 112, 6313–6318 (2015).
Reinhardt, A. & Frenkel, D. Numerical evidence for nucleated self-assembly of DNA brick structures. Phys. Rev. Lett. 112, 238103 (2014).
Hormoz, S. & Brenner, M. P. Design principles for self-assembly with short-range interactions. Proc. Natl Acad. Sci. USA 108, 5193–5198 (2011).
Zeravcic, Z., Manoharan, V. N. & Brenner, M. P. Size limits of self-assembled colloidal structures made using specific interactions. Proc. Natl Acad. Sci. USA 111, 15918–15923 (2014).
Tkachenko, A. V. Theory of programmable hierarchic self-assembly. Phys. Rev. Lett. 106, 255501 (2011).
Halverson, J. D. & Tkachenko, A. V. DNA-programmed mesoscopic architecture. Phys. Rev. E 87, 062310 (2013).
Wu, K.-T. et al. Polygamous particles. Proc. Natl Acad. Sci. USA 109, 18731–18736 (2012).
Wang, Y. et al. Colloids with valence and specific directional bonding. Nature 491, 51–55 (2012). This paper describes the synthesis and assembly of colloidal microparticles with both specific and directional interactions; such particles might be able to assemble like DNA bricks if a sufficient number of different species can be synthesized.
Feng, L., Dreyfus, R., Sha, R., Seeman, N. C. & Chaikin, P. M. DNA patchy particles. Adv. Mater. 25, 2779–2783 (2013).
Maye, M. M., Nykypanchuk, D., Cuisinier, M., van der Lelie, D. & Gang, O. Stepwise surface encoding for high-throughput assembly of nanoclusters. Nat. Mater. 8, 388–391 (2009).
Jones, M. R. et al. DNA-nanoparticle superlattices formed from anisotropic building blocks. Nat. Mater. 9, 913–917 (2010).
Lu, F., Yager, K. G., Zhang, Y., Xin, H. & Gang, O. Superlattices assembled through shape-induced directional binding. Nat. Commun. 6, 6912 (2015).
O’Brien, M. N., Jones, M. R., Lee, B. & Mirkin, C. A. Anisotropic nanoparticle complementarity in DNA-mediated co-crystallization. Nat. Mater. 14, 833–839 (2015).
Woo, S. & Rothemund, P. W. K. Programmable molecular recognition based on the geometry of DNA nanostructures. Nat. Chem. 3, 620–627 (2011).
Endo, M., Sugita, T., Katsuda, Y., Hidaka, K. & Sugiyama, H. Programmed-assembly system using DNA jigsaw pieces. Chem. Eur. J. 16, 5362–5368 (2010).
Rajendran, A., Endo, M., Katsuda, Y., Hidaka, K. & Sugiyama, H. Programmed two-dimensional self-assembly of multiple DNA origami jigsaw pieces. ACS Nano 5, 665–671 (2011).
Gerling, T., Wagenbauer, K. F., Neuner, A. M. & Dietz, H. Dynamic DNA devices and assemblies formed by shape-complementary, non-base pairing 3D components. Science 347, 1446–1452 (2015).
Tian, Y. et al. Prescribed nanoparticle cluster architectures and low-dimensional arrays built using octahedral DNA origami frames. Nat. Nanotechnol. 10, 637–644 (2015). Experimental study showing that DNA origami can be used to organize the assembly of nanoparticles, demonstrating what can be achieved by combining structural DNA nanotechnology with DNA-grafted nanoparticles.
Roldán-Vargas, S., Smallenburg, F., Kob, W. & Sciortino, F. Gelling by heating. Sci. Rep. 3, 2451 (2013).
Rogers, W. B. & Manoharan, V. N. Programming colloidal phase transitions with DNA strand displacement. Science 347, 639–642 (2015).
Zhang, D. Y. & Winfree, E. Control of DNA strand displacement kinetics using toehold exchange. J. Am. Chem. Soc. 131, 17303–17314 (2009).
Golestanian, R., Liverpool, T. B. & Ajdari, A. Propulsion of a molecular machine by asymmetric distribution of reaction products. Phys. Rev. Lett. 94, 220801 (2005).
Wang, W., Duan, W., Ahmed, S., Mallouk, T. E. & Sen, A. Small power: autonomous nano- and micromotors propelled by self-generated gradients. Nano Today 8, 531–554 (2013).
Keber, F. C. et al. Topology and dynamics of active nematic vesicles. Science 345, 1135–1139 (2014).
Yurke, B., Turberfield, A. J., Mills, A. P. Jr, Simmel, F. C. & Neumann, J. L. A DNA-fuelled molecular machine made of DNA. Nature 406, 605–608 (2000).
Winfree, E. & Bekbolatov, R. in DNA Computing (eds Chen, J.& Reif, J.) 126–144 (Springer, 2003).
Murugan, A., Huse, D. A. & Leibler, S. Discriminatory proofreading regimes in nonequilibrium systems. Phys. Rev. X 4, 021016 (2014).
Shin, J.-S. & Pierce, N. A. A synthetic DNA walker for molecular transport. J. Am. Chem. Soc. 126, 10834–10835 (2004).
Lund, K. et al. Molecular robots guided by prescriptive landscapes. Nature 465, 206–210 (2010).
Leunissen, M. E. et al. Towards self-replicating materials of DNA-functionalized colloids. Soft Matter 5, 2422–2430 (2009).
Wang, T. et al. Self-replication of information-bearing nanoscale patterns. Nature 478, 225–228 (2011).
Zeravcic, Z. & Brenner, M. P. Self-replicating colloidal clusters. Proc. Natl Acad. Sci. USA 111, 1748–1753 (2014).
Li, C.-J. & Trost, B. M. Green chemistry for chemical synthesis. Proc. Natl Acad. Sci. USA 105, 13197–13202 (2008).
Kosuri, S. & Church, G. M. Large-scale de novo DNA synthesis: technologies and applications. Nat. Methods 11, 499–507 (2014).
Zhang, Y. et al. Selective transformations between nanoparticle superlattices via the reprogramming of DNA-mediated interactions. Nat. Mater. 14, 840–847 (2015).
Acknowledgements
We thank M. Brenner, D. Frenkel, O. Gang, A. Tkachenko, W. Jacobs, D. Pine, P. Chaikin, J. Crocker and B. Mognetti for discussions and the National Science Foundation for funding through grant no. DMR-1435964.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Rights and permissions
About this article
Cite this article
Rogers, W., Shih, W. & Manoharan, V. Using DNA to program the self-assembly of colloidal nanoparticles and microparticles. Nat Rev Mater 1, 16008 (2016). https://doi.org/10.1038/natrevmats.2016.8
Published:
DOI: https://doi.org/10.1038/natrevmats.2016.8
This article is cited by
-
Macroscopic photonic single crystals via seeded growth of DNA-coated colloids
Nature Communications (2023)
-
Functionalization and higher-order organization of liposomes with DNA nanostructures
Nature Communications (2023)
-
Long-Range Ordered Nanostructures of Assembling Macromolecules via Rational Design of Kinetic Pathways: A Computational Perspective
Chinese Journal of Polymer Science (2023)
-
Comprehensive view of microscopic interactions between DNA-coated colloids
Nature Communications (2022)
-
Self-assembly of emulsion droplets through programmable folding
Nature (2022)