Abstract
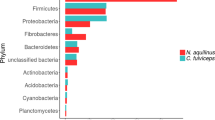
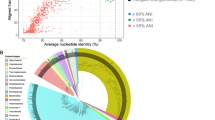
From the standpoints of both basic research and biotechnology, there is considerable interest in reaching a clearer understanding of the diversity of biological mechanisms employed during lignocellulose degradation. Globally, termites are an extremely successful group of wood-degrading organisms1 and are therefore important both for their roles in carbon turnover in the environment and as potential sources of biochemical catalysts for efforts aimed at converting wood into biofuels. Only recently have data supported any direct role for the symbiotic bacteria in the gut of the termite in cellulose and xylan hydrolysis2. Here we use a metagenomic analysis of the bacterial community resident in the hindgut paunch of a wood-feeding ‘higher’ Nasutitermes species (which do not contain cellulose-fermenting protozoa) to show the presence of a large, diverse set of bacterial genes for cellulose and xylan hydrolysis. Many of these genes were expressed in vivo or had cellulase activity in vitro, and further analyses implicate spirochete and fibrobacter species in gut lignocellulose degradation. New insights into other important symbiotic functions including H2 metabolism, CO2-reductive acetogenesis and N2 fixation are also provided by this first system-wide gene analysis of a microbial community specialized towards plant lignocellulose degradation. Our results underscore how complex even a 1-μl environment can be.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Sugimoto, A., Bignell, D. E. & Macdonald, J. A. in Termites: Evolution, Sociality, Symbioses, Ecology (eds Abe, T. Bignell, D. E. & Higashi, M.) 409–435 (Kluwer Academic, Dordrecht, 2000)
Tokuda, G. & Watanabe, H. Hidden cellulases in termites: revision of an old hypothesis. Biol. Lett. 3, 336–339 (2007)
Brune, A. in The Prokaryotes Vol. 1 (eds Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H. & Stackebrandt, E.) 439–474 (Springer, New York, 2006)
Kambhampati, S. & Eggleton, P. in Termites: Evolution, Sociality, Symbioses, Ecology (eds Abe, T. Bignell, D. E. & Higashi, M.) 1–24 (Kluwer Academic, Dordrecht, 2000)
Tokuda, G. et al. Major alteration of the expression site of endogenous cellulases in members of an apical termite lineage. Mol. Ecol. 13, 3219–3228 (2004)
Brennan, Y. et al. Unusual microbial xylanases from insect guts. Appl. Environ. Microbiol. 70, 3609–3617 (2004)
McHardy, A. C., Martin, H. G., Tsirigos, A., Hugenholtz, P. & Rigoutsos, I. Accurate phylogenetic classification of variable-length DNA fragments. Nat. Methods 4, 63–72 (2007)
Brune, A., Emerson, D. & Breznak, J. A. The termite gut microflora as an oxygen sink: microelectrode determination of oxygen and pH gradients in guts of lower and higher termites. Appl. Environ. Microbiol. 61, 2681–2687 (1995)
Davies, G. J. & Henrissat, B. Structural enzymology of carbohydrate-active enzymes: implications for the post-genomic era. Biochem. Soc. Trans. 30, 291–297 (2002)
Breznak, J. A. & Brune, A. Role of microorganisms in the digestion of lignocellulose in termites. Annu. Rev. Entomol. 39, 453–487 (1994)
Taylor, L. E. et al. Complete cellulase system in the marine bacterium Saccharophagus degradans strain 2-40T. J. Bacteriol. 188, 3849–3861 (2006)
Xie, G. et al. Genome sequence of the cellulolytic gliding bacterium Cytophaga hutchinsonii . Appl. Environ. Microbiol. 73, 3536–3546 (2007)
Tringe, S. G. et al. Comparative metagenomics of microbial communities. Science 308, 554–557 (2005)
Venter, J. C. et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304, 66–74 (2004)
Gill, S. R. et al. Metagenomic analysis of the human distal gut microbiome. Science 312, 1355–1359 (2006)
Qi, M. et al. Novel molecular features of the fibrolytic intestinal bacterium Fibrobacter intestinalis not shared with Fibrobacter succinogenes as determined by suppressive subtractive hybridization. J. Bacteriol. 187, 3739–3751 (2005)
Odelson, D. A. & Breznak, J. A. Nutrition and growth characteristics of Trichomitopsis termopsidis, a cellulolytic protozoan from termites. Appl. Environ. Microbiol. 49, 614–621 (1985)
Vignais, P. M. & Colbeau, A. Molecular biology of microbial hydrogenases. Curr. Issues Mol. Biol. 6, 159–188 (2004)
Breznak, J. A. & Switzer, J. M. Acetate synthesis from H2 plus CO2 by termite gut microbes. Appl. Environ. Microbiol. 52, 623–630 (1986)
Pester, M. & Brune, A. Hydrogen is the central free intermediate during lignocellulose degradation by termite gut symbionts. ISME J. 1, 551–565 (2007)
Ragsdale, S. W. The eastern and western branches of the Wood/Ljungdahl pathway: how the East and West were won. Biofactors 6, 3–11 (1997)
Pester, M. & Brune, A. Expression profiles of fhs (FTHFS) genes support the hypothesis that spirochaetes dominate reductive acetogenesis in the hindgut of lower termites. Environ. Microbiol. 8, 1261–1270 (2006)
Prestwich, G. D., Bentley, B. L. & Carpenter, E. J. Nitrogen sources for neotropical nasute termites: Fixation and selective foraging. Oecologia 46, 397–401 (1980)
Ohkuma, M., Noda, S. & Kudo, T. Phylogenetic diversity of nitrogen fixation genes in the symbiotic microbial community in the gut of diverse termites. Appl. Environ. Microbiol. 65, 4926–4934 (1999)
Potrikus, C. J. & Breznak, J. A. Anaerobic degradation of uric acid by gut bacteria of termites. Appl. Environ. Microbiol. 40, 125–132 (1980)
Brune, A. & Friedrich, M. Microecology of the termite gut: structure and function on a microscale. Curr. Opin. Microbiol. 3, 263–269 (2000)
Beckwith, T. & Light, S. The spirals within the termite gut for class use. Science 66, 656–657 (1927)
Robertson, D. E. et al. Exploring nitrilase sequence space for enantioselective catalysis. Appl. Environ. Microbiol. 70, 2429–2436 (2004)
Short, J. M., Fernandez, J. M., Sorge, J. A. & Huse, W. D. Lambda ZAP: a bacteriophage lambda expression vector with in vivo excision properties. Nucleic Acids Res. 16, 7583–7600 (1988)
Ewing, B. & Green, P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8, 186–194 (1998)
Ewing, B., Hillier, L., Wendl, M. C. & Green, P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8, 175–185 (1998)
Markowitz, V. M. et al. An experimental metagenome data management and analysis system. Bioinformatics 22, e359–e367 (2006)
Markowitz, V. M. et al. The integrated microbial genomes (IMG) system. Nucleic Acids Res. 34, D344–D348 (2006)
Marchler-Bauer, A., Panchenko, A. R., Ariel, N. & Bryant, S. H. Comparison of sequence and structure alignments for protein domains. Proteins 48, 439–446 (2002)
Emanuelsson, O., Brunak, S., von Heijne, G. & Nielsen, H. Locating proteins in the cell using TargetP, SignalP and related tools. Nature Protocols 2 953–971 doi: 10.1038/nprot.2007.131 (2007)
Wei, J. et al. Global proteome discovery using an online three-dimensional LC-MS/MS. J. Proteome Res. 4, 801–808 (2005)
DeSantis, T. Z. et al. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 34, W394–W399 (2006)
DeSantis, T. Z. et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072 (2006)
Stamatakis, A., Ludwig, T. & Meier, H. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics 21, 456–463 (2005)
Schmidt, H. A., Strimmer, K., Vingron, M. & von Haeseler, A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18, 502–504 (2002)
Schloss, P. D. & Handelsman, J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71, 1501–1506 (2005)
Colwell, R. K. & Coddington, J. A. Estimating terrestrial biodiversity through extrapolation. Phil. Trans. R. Soc. Lond. B 345, 101–118 (1994)
de Caprariis, P., Lindemann, R. & Collins, C. A method for determining optimum sample size in species diversity studies. J. Int. Assoc. Math. Geol. 8, 575–581 (1976)
Smith, E. P. & van Belle, G. Nonparametric estimation of species richness. Biometrics 40, 119–129 (1984)
Krebs, C. J. Ecological Methodology (Harper & Row, New York, 1989)
Efron, B. & Tibshirani, R. An Introduction to the Bootstrap (Chapman & Hall, New York, 1993)
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004)
Ludwig, W. et al. ARB: a software environment for sequence data. Nucleic Acids Res. 32, 1363–1371 (2004)
Austin, J. W., Szalanski, A. L. & Cabrera, B. J. Phylogenetic analysis of the subterranean termite family Rhinotermitidae (Isoptera) by using the mitochondrial cytochrome oxidase II gene. Ann. Entomol. Soc. Am. 97, 548–555 (2004)
Ohkuma, M. et al. Molecular phylogeny of Asian termites (Isoptera) of the families Termitidae and Rhinotermitidae based on mitochondrial COII sequences. Mol. Phylogenet. Evol. 31, 701–710 (2004)
Henrissat, B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280, 309–316 (1991)
Davies, D. G. & Geesey, G. G. Regulation of the alginate biosynthesis gene algC in Pseudomonas aeruginosa during biofilm development in continous culture. Appl. Environ. Microbiol. 61, 860–867 (1995)
Johnston, D. in Handbook of Food Enzymology (eds Whitaker, J. R., Voragen, A. G. J. & Wong, D. W. S.) 761–769 (Marcel Dekker Inc., New York, 2002)
Stahlberg, J., Johansson, G. & Pettersson, G. Trichoderma reesei has no true exo-cellulase: all intact and truncated cellulases produce new reducing end groups on cellulose. Biochim. Biophys. Acta 1157, 107–113 (1993)
Wood, T. Preparation of crystalline, amorphous and dyed cellulase substrate. Methods Enzymol. 160, 19–25 (1988)
von Mering, C. et al. STRING 7—recent developments in the integration and prediction of protein interactions. Nucleic Acids Res. 35, D358–D362 (2007)
Marchler-Bauer, A. & Bryant, S. H. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32, W327–W331 (2004)
Marchler-Bauer, A. et al. CDD: a conserved domain database for protein classification. Nucleic Acids Res. 33, D192–D196 (2005)
Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16, 10881–10890 (1988)
Apweiler, R. et al. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 29, 37–40 (2001)
Peters, J. W., Lanzilotta, W. N., Lemon, B. J. & Seefeldt, L. C. X-ray crystal structure of the Fe-only hydrogenase (CpI) from Clostridium pasteurianum to 1.8 angstrom resolution. Science 282, 1853–1858 (1998)
Chenna, R. et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31, 3497–3500 (2003)
Bates, P. A., Kelley, L. A., MacCallum, R. M. & Sternberg, M. J. E. Enhancement of protein modeling by human intervention in applying the automatic programs 3D-JIGSAW and 3D-PSSM. Proteins Struct. Funct. Genet. 45, 39–46 (2001)
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 27–28 (1996)
Lilburn, T. G. et al. Nitrogen fixation by symbiotic and free-living spirochetes. Science 292, 2495–2498 (2001)
Sturn, A., Quackenbush, J. & Trajanoski, Z. Genesis: cluster analysis of microarray data. Bioinformatics 18, 207–208 (2002)
Acknowledgements
We thank production and sequencing teams at Verenium and the Joint Genome Institute for their expertise, J. Mata for morphological identifications, L. Christoffersen for logistic and permitting support, and our laboratory colleagues for their comments during manuscript preparation. This research was supported in part by the National Science Foundation, the National Institutes of Health, Caltech, and the Lawrence Berkeley National Laboratory. The sequencing for the project was provided through the US Department of Energy (DOE) Community Sequencing Program (http://www.jgi.doe.gov/CSP/index.html). This work was performed, in part, under the auspices of the DOE Office of Science, Biological and Environmental Research Program, University of California, Lawrence Livermore National Laboratory, and Los Alamos National Laboratory.
Author Contributions F.W., P.H., E.J.M., D.E.R., E.M.R. and J.R.L. performed project planning, coordination, execution and facilitation. M.H., C.M., L.G.A. and G.T. undertook field research planning, permits, logistics and station management. F.W., M.C., M.H., C.M., L.G.A. and J.R.L. conducted field collection and sample preparation. M.C., G.D., N.A., J.M. and C.C. performed nucleic acid purification and library construction. D.P. and K.B. carried out assemblies. A.S. conducted gene prediction and annotation. E.S. undertook data processing and loading into IMG/M. N.I. and N.C.K. performed metabolic reconstruction. A.C.M. and I.R. carried out binning. N.M. conducted fosmid annotation and manual curation. M.G., J.T.S. and B.D.G. performed proteomics and enzyme activities. F.W., P.L., V.K., D.D., E.K., E.G.M., E.A.O. and X.Z. carried out phylogenetic analyses. H.G.M. made accumulation curves, diversity estimates, statistical test for gene-centric analysis. P.L., T.H.R. and J.R.L. performed glycoside hydrolase bioinformatics. R.S. and S.G.T. constructed hypothetical gene families. N.I. and P.H. were responsible for hydrogenases. E.G.M., E.A.O., X.Z. and J.R.L. performed C1-pathway and N-metabolic reconstruction. M.P. carried out sensory transduction protein analysis. F.W., P.L., M.G., T.H.R., J.T.S., P.H., N.I., R.S., S.G.T., M.P. and J.R.L. undertook manuscript preparation.
This whole-genome shotgun project has been deposited at DDBJ/EMBL/GenBank under accession number ABDH00000000; this first version is ABDH01000000. The COII gene from the termite host is deposited under accession number EU236539, and 16S rRNA gene sequences are deposited under the accession numbers EF453758–EF455009 and EU024891–EU024927. The subcloned cellulase gene sequences are deposited under the accession numbers EF428062–EF428109.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Several authors are affiliated with businesses with commercial interests in lignocellulose decay. Verenium Corporation has commercial interests in enzymes associated with lignocellulose decay.
Supplementary information
Supplementary Information
The file contains Supplementary Materials and Methods, Supplementary Tables S1-S11 and Supplementary Figures S1-S23 with Legends. (PDF 10773 kb)
Rights and permissions
About this article
Cite this article
Warnecke, F., Luginbühl, P., Ivanova, N. et al. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature 450, 560–565 (2007). https://doi.org/10.1038/nature06269
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature06269
This article is cited by
-
Metagenomic analysis of gut microbiome illuminates the mechanisms and evolution of lignocellulose degradation in mangrove herbivorous crabs
BMC Microbiology (2024)
-
Gut microbiota variations in wild yellow baboons (Papio cynocephalus) are associated with sex and habitat disturbance
Scientific Reports (2024)
-
The gut microbiota of insects: a potential source of bacteria and metabolites
International Journal of Tropical Insect Science (2024)
-
Impact of gut microbiota composition on black cutworm, Agrotis ipsilon (hufnagel) metabolic indices and pesticide degradation
Animal Microbiome (2023)
-
Exploring the microbial diversity and characterization of cellulase and hemicellulase genes in goat rumen: a metagenomic approach
BMC Biotechnology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.