Abstract
Collective movement of epithelial cells drives essential multicellular organization during various fundamental physiological processes encompassing embryonic morphogenesis, cancer and wound healing. Yet the molecular mechanism that ensures the coordinated movement of many cells remains elusive. Here we show that a tumour suppressor protein, merlin, coordinates collective migration of tens of cells, by acting as a mechanochemical transducer. In a stationary epithelial monolayer and also in three-dimensional human skin, merlin localizes to cortical cell–cell junctions. During migration initiation, a fraction of cortical merlin relocalizes to the cytoplasm. This relocalization is triggered by the intercellular pulling force of the leading cell and depends on the actomyosin-based cell contractility. Then in migrating cells, taking its cue from the intercellular pulling forces, which show long-distance ordering, merlin coordinates polarized Rac1 activation and lamellipodium formation on the multicellular length scale. Together, these results provide a distinct molecular mechanism linking intercellular forces to collective cell movements in migrating epithelia.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Rorth, P. Fellow travellers: emergent properties of collective cell migration. EMBO Rep. 13, 984–991 (2012).
Friedl, P. & Gilmour, D. Collective cell migration in morphogenesis, regeneration and cancer. Nat. Rev. Mol. Cell Biol. 10, 445–457 (2009).
Friedl, P., Locker, J., Sahai, E. & Segall, J. E. Classifying collective cancer cell invasion. Nat. Cell Biol. 14, 777–783 (2012).
Ramel, D., Wang, X. B., Laflamme, C., Montell, D. J. & Emery, G. Rab11 regulates cell–cell communication during collective cell movements. Nat. Cell Biol. 15, 317–324 (2013).
Theveneau, E. et al. Chase-and-run between adjacent cell populations promotes directional collective migration. Nat. Cell Biol. 15, 763 (2013).
Simpson, K. J. et al. Identification of genes that regulate epithelial cell migration using an siRNA screening approach. Nat. Cell Biol. 10, 1027–1038 (2008).
Poujade, M. et al. Collective migration of an epithelial monolayer in response to a model wound. Proc. Natl Acad. Sci. USA 104, 15988–15993 (2007).
Angelini, T. E., Hannezo, E., Trepat, X., Fredberg, J. J. & Weitz, D. A. Cell Migration Driven by Cooperative Substrate Deformation Patterns. Phys. Rev. Lett. 104, 168104 (2010).
Petitjean, L. et al. Velocity Fields in a Collectively Migrating Epithelium. Biophys. J. 98, 1790–1800 (2010).
Gaggioli, C. et al. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat. Cell Biol. 9, 1392–1400 (2007).
Xu, N., Keung, B. & Myat, M. M. Rho GTPase controls invagination and cohesive migration of the Drosophila salivary gland through Crumbs and Rho-kinase. Dev. Biol. 321, 88–100 (2008).
Wang, X. B., He, L., Wu, Y. I., Hahn, K. M. & Montell, D. J. Light-mediated activation reveals a key role for Rac in collective guidance of cell movement in vivo. Nat. Cell. Biol. 12, 591–597 (2010).
Hidalgo-Carcedo, C. et al. Collective cell migration requires suppression of actomyosin at cell–cell contacts mediated by DDR1 and the cell polarity regulators Par3 and Par6. Nat. Cell Biol. 13, 49–58 (2011).
Ng, M. R., Besser, A., Danuser, G. & Brugge, J. S. Substrate stiffness regulates cadherin-dependent collective migration through myosin-II contractility. J. Cell Biol. 199, 545–563 (2012).
Reffay, M. et al. Interplay of RhoA and mechanical forces in collective cell migration driven by leader cells. Nat. Cell Biol. 16, 217–223 (2014).
Peglion, F., Llense, F. & Etienne-Manneville, S. Adherens junction treadmilling during collective migration. Nat. Cell Biol. 16, 639–651 (2014).
Tambe, D. T. et al. Collective cell guidance by cooperative intercellular forces. Nat. Mater. 10, 469–475 (2011).
Trepat, X. et al. Physical forces during collective cell migration. Nat. Phys. 5, 426–430 (2009).
Angelini, T. E. et al. Glass-like dynamics of collective cell migration. Proc. Natl Acad. Sci. USA 108, 4714–4719 (2011).
Dupont, S. et al. Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183 (2011).
Bernascone, I. & Martin-Belmonte, F. Crossroads of Wnt and Hippo in epithelial tissues. Trends Cell Biol. 23, 380–389 (2013).
Bosveld, F. et al. Mechanical control of morphogenesis by Fat/Dachsous/Four-Jointed planar cell polarity pathway. Science 336, 724–727 (2012).
Fehon, R. G., McClatchey, A. I. & Bretscher, A. Organizing the cell cortex: the role of ERM proteins. Nat. Rev. Mol. Cell Biol. 11, 276–287 (2010).
Hamaratoglu, F. et al. The tumour-suppressor genes NF2/Merlin and expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat. Cell Biol. 8, 27–36 (2006).
Li, W., Cooper, J., Karajannis, M. A. & Giancotti, F. G. Merlin: a tumour suppressor with functions at the cell cortex and in the nucleus. EMBO Rep. 13, 204–215 (2012).
Cole, B. K., Curto, M., Chan, A. W. & McClatchey, A. I. Localization to the cortical cytoskeleton is necessary for Nf2/Merlin-dependent epidermal growth factor receptor silencing. Mol. Cell Biol. 28, 1274–1284 (2008).
Wozniak, M. A. & Chen, C. S. Mechanotransduction in development: a growing role for contractility. Nat. Rev. Mol. Cell Biol. 10, 34–43 (2009).
Vogel, V. & Sheetz, M. Local force and geometry sensing regulate cell functions. Nat. Rev. Mol. Cell Biol. 7, 265–275 (2006).
Tse, J. M. et al. Mechanical compression drives cancer cells toward invasive phenotype. Proc. Natl Acad. Sci. USA 109, 911–916 (2012).
Serra-Picamal, X. et al. Mechanical waves during tissue expansion. Nat. Phys. 8, 628–634 (2012).
Yi, C. L. et al. A Tight junction-associated merlin-angiomotin complex Mediates Merlin’s regulation of mitogenic signaling and tumor suppressive functions. Cancer Cell 19, 527–540 (2011).
Lallemand, D., Saint-Amaux, A. L. & Giovannini, M. Tumor-suppression functions of merlin are independent of its role as an organizer of the actin cytoskeleton in Schwann cells. J. Cell Sci. 122, 4141–4149 (2009).
Machacek, M. et al. Coordination of Rho GTPase activities during cell protrusion. Nature 461, 99–103 (2009).
Wu, Y. I. et al. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature 461, 104–108 (2009).
Fenteany, G., Janmey, P. A. & Stossel, T. P. Signaling pathways and cell mechanics involved in wound closure by epithelial cell sheets. Curr. Biol. 10, 831–838 (2000).
Okada, T., Lopez-Lago, M. & Giancotti, F. G. Merlin/NF-2 mediates contact inhibition of growth by suppressing recruitment of Rac to the plasma membrane. J. Cell Biol. 171, 361–371 (2005).
Guo, L., Moon, C., Niehaus, K., Zheng, Y. & Ratner, N. Rac1 Controls Schwann Cell Myelination through cAMP and NF2/merlin. J. Neurosci. 32, 17251–17261 (2012).
Gladden, A. B., Hebert, A. M., Schneeberger, E. E. & McClatchey, A. I. The NF2 tumor suppressor, Merlin, regulates epidermal development through the establishment of a junctional polarity complex. Dev. Cell 19, 727–739 (2010).
Hebert, A. M., DuBoff, B., Casaletto, J. B., Gladden, A. B. & McClatchey, A. I. Merlin/ERM proteins establish cortical asymmetry and centrosome position. Genes Dev. 26, 2709–2723 (2012).
Flaiz, C., Kaempchen, K., Matthies, C. & Hanemann, C. O. Actin-rich protrusions and nonlocalized GTPase activation in Merlin-deficient schwannomas. J. Neuropathol. Exp. Neurol. 66, 608–616 (2007).
Shaw, R. J. et al. The Nf2 tumor suppressor, merlin, functions in Rac-dependent signaling. Dev. Cell 1, 63–72 (2001).
Farooqui, R. & Fenteany, G. Multiple rows of cells behind an epithelial wound edge extend cryptic lamellipodia to collectively drive cell-sheet movement. J. Cell Sci. 118, 51–63 (2005).
Schubert, R. A., Dodd, I. B., Egan, J. B. & Shearwin, K. E. Cro’s role in the CI-Cro bistable switch is critical for lambda’s transition from lysogeny to lytic development. Genes Dev. 21, 2461–2472 (2007).
Huang, B. et al. The three-way switch operation of Rac1/RhoA GTPase-based circuit controlling amoeboid-hybrid-mesenchymal transition. Sci. Rep. 4, 6449 (2014).
Saito, K., Ozawa, Y., Hibino, K. & Ohta, Y. FilGAP, a Rho/Rho-associated protein kinase-regulated GTPase-activating protein for Rac, controls tumor cell migration. Mol. Biol. Cell 23, 4739–4750 (2012).
Sanz-Moreno, V. et al. Rac activation and inactivation control plasticity of tumor cell movement. Cell 135, 510–523 (2008).
Alfthan, K., Heiska, L., Gronholm, M., Renkema, G. H. & Carpen, O. Cyclic AMP-dependent protein kinase phosphorylates merlin at serine 518 independently of p21-activated kinase and promotes merlin-ezrin heterodimerization. J. Biol. Chem. 279, 18559–18566 (2004).
Jin, H. C., Sperka, T., Herrlich, P. & Morrison, H. Tumorigenic transformation by CPI-17 through inhibition of a merlin phosphatase. Nature 442, 576–579 (2006).
Safferling, K. et al. Wound healing revised: a novel reepithelialization mechanism revealed by in vitro and in silico models. J. Cell Biol. 203, 691–709 (2013).
Trepat, X. & Fredberg, J. J. Plithotaxis and emergent dynamics in collective cellular migration. Trends Cell Biol. 21, 638–646 (2011).
Lukeman, R., Li, Y. X. & Edelstein-Keshet, L. Inferring individual rules from collective behavior. Proc. Natl Acad. Sci. USA 107, 12576–12580 (2010).
Sumino, Y. et al. Large-scale vortex lattice emerging from collectively moving microtubules. Nature 483, 448–452 (2012).
Yates, C. A. et al. Inherent noise can facilitate coherence in collective swarm motion. Proc. Natl Acad. Sci. USA 106, 5464–5469 (2009).
Fang, X. F. et al. The SOX2 response program in glioblastoma multiforme: an integrated ChIP-seq, expression microarray, and microRNA analysis. BMC Genomics 12, 11 (2011).
Dang, I. et al. Inhibitory signalling to the Arp2/3 complex steers cell migration. Nature 503, 281–284 (2013).
Jungbauer, S., Gao, H. J., Spatz, J. P. & Kemkemer, R. Two characteristic regimes in frequency-dependent dynamic reorientation of fibroblasts on cyclically stretched substrates. Biophys. J. 95, 3470–3478 (2008).
Harris, A. R. & Charras, G. T. Experimental validation of atomic force microscopy-based cell elasticity measurements. Nanotechnology 22, 345102 (2011).
Grashoff, C. et al. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature 466, 263–266 (2010).
Feder, T. J., BrustMascher, I., Slattery, J. P., Baird, B. & Webb, W. W. Constrained diffusion or immobile fraction on cell surfaces: a new interpretation. Biophys. J. 70, 2767–2773 (1996).
Das, T., Maiti, T. K. & Chakraborty, S. Augmented stress-responsive characteristics of cell lines in narrow confinements. Integr. Biol. 3, 684–695 (2011).
Rausch, S. et al. Polarizing cytoskeletal tension to induce leader cell formation during collective cell migration. Biointerphases 8, 32 (2013).
Tseng, Q. et al. Spatial organization of the extracellular matrix regulates cell–cell junction positioning. Proc. Natl Acad. Sci. USA 109, 1506–1511 (2012).
Tambe, D. T. et al. Monolayer stress microscopy: limitations, artifacts, and accuracy of recovered intercellular stresses. PLoS ONE 8, e55172 (2013).
Acknowledgements
We are grateful to B. Geiger and F. Frischknecht for critical review of the manuscript and advice. This work was supported by the Max Planck Society. J.P.S. is the Weston Visiting Professor at the Weizmann Institute of Science and is a member of the Heidelberg Cluster of Excellence CellNetworks. This work was supported by CellNetworks EcTop2 and the Grassroot Project of the Max Planck Institute for Intelligent Systems. Part of this work was supported by the BMBF/MPG network MaxSynBio.
Author information
Authors and Affiliations
Contributions
T.D., H.B. and J.P.S. conceived the project, analysed the data and wrote the manuscript. T.D. carried out all experiments, except those experiments that involve wound healing in the 3D skin model, which were carried out by K.S. and N.G. S.R. provided technical help. All authors went through the manuscript and provided feedback.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 4 Effect of Merlin depletion on velocity correlation function and correlations lengths.
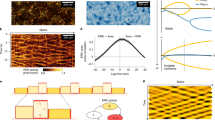
(a) Representative images showing the sheet migration of MDCK monolayer at the indicated times after the removal of confinement. Scale bars, 100 μm. (b–c) Time-lapse phase-contrast images (b) of migrating epithelial monolayer were analysed by PIV to determine the velocity vectors (c). Scale bars, 150 μm. Magnified panel (c, Right, Scale bar, 50 μm) shows a highly correlated velocity domain. (d) Lateral and axial velocity correlation functions were calculated as indicated by the expressions. (e) Representative lateral (Left) and axial (Right) velocity correlation function. Correlation length is the distance where correlation function becomes zero. (f) (Left) While lateral correlation function in DECMA-1 treated cells (cells with reduced intercellular connection; green squares) showed a drastic decay in comparison to control cells (red circles), (Right) no such trend was observed in axial correlation function. (g) Lateral and axial velocity correlation lengths of DECMA-1 treated and control cells. (n = 15 samples from 3 independent experiments in all cases; Lateral control versus DECMA-1, P = 7.79 × 10−26; Axial control versus DECMA-1, P = 0.51; Lateral DECMA-1 versus axial DECMA-1, P = 1.49 × 10−12; Wilcoxon rank-sum test) ∗∗∗P < 0.001, NS: not significant (Wilcoxon rank-sum test). In box plots central mark is the median, and the edges of the box are the 1st and 3rd quartiles. Whiskers extend to the most extreme non-outlier data points. (h) Synopsis of the results from the screening experiments in HaCaT cells. Values written in the color-coded table are the mean correlation length. n = 6 samples from 3 independent experiments performed with each siRNA (Supplementary Table 1). Lack of response due to failure in gene knockdown cannot be ruled out. (i) Comparison of the representative lateral velocity correlation functions in Merlin-depleted versus scramble siRNA-transfected (control) MDCK cells showed a drastic decrease of correlation in Merlin-depleted cells. (j) Color map of velocity vectors revealed highly correlated velocity domains (indicated with arrow) in scramble siRNA-transfected MDCK cells. (k) In contrast, no such domain could be observed in Merlin-depleted cells. In (j, k), scale bars, 100 μm. Panels (a–c, e,f, i-k) show the representative image of 3 independent experiments.
Supplementary Figure 5 Effect of Merlin-depletion on cell migration and integrity of cell–cell junctions.
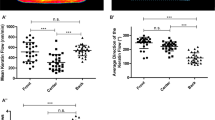
(a) Representative migration tracks of cells, transfected either with scramble siRNA (Left) or with Merlin siRNA1 (Right). (b–d) Directional persistence of cell migration (b, computed as the ratio of the distance between two points by the actual trajectory), individual cell speed (c), and collective speed (d) in control and Merlin-depleted MDCK monolayers. In (b) and (c), n = number of cells from 3 independent experiments (mean ± s.e.m; b, P = 3.55 × 10−57; c, P = 4.66 × 10−21; Wilcoxon rank-sum test). In (d), n = number of samples from 3 independent experiments (mean ± s.e.m; P = 3.84 × 10−16; Wilcoxon rank-sum test). (e) Mean individual cell speed in confluent control and Merlin-depleted MDCK cell monolayer. (n = 5 samples from 5 independent experiments in all cases, P = 0.07; Wilcoxon rank-sum test) For each sample, 20 or more cells were counted. In box plots central mark is the median, and the edges of the box are the 1st and 3rd quartiles. Whiskers extend to the most extreme non-outlier data points. (f) Phase-contrast image (Left) and traction force landscape (Right) of a MDCK monolayer moving towards right side. Scale bars, 100 μm. (g) Distribution of the axial (w.r.t. the direction of collective migration) traction forces showed broad-tailed distribution in both control (green circles) and Merlin-depleted (red squares) cells. (h) Adherens and tight junctions as marked by E-cadherin and ZO-1, respectively, in control and Merlin-depleted MDCK cells. Scale bars, 20 μm. (i) Z-section of confocal images showing the distribution of E-cadherin and ZO-1. Scale bars, 10 μm. (j) FRAP curve showing E-cadherin recovery after photobleaching in E-cadherin-GFP transfected control MDCK cells. t1/2 is the time required to attain 50% of the final recovered intensity (mobile fraction). n = 10 independent experiments; mean ± s.e.m. (k, l) Immobile fraction (k) and t1/2 (l) in control and Merlin siRNA1 transfected cells (by either forward or reverse transfection method). In (k,l), n = 50 cells from 3 independent experiments in all cases (mean ± s.e.m; k, Control versus Forward, P = 0.53; Control versus Reverse, P = 0.08; l, Control versus Forward, P = 0.65; Control versus Reverse, P = 0.28; Wilcoxon rank-sum test). ∗∗∗P < 0.001, NS: not significant (Wilcoxon rank-sum test). Panels (a, f, i) show the representative image of 3 independent experiments.
Supplementary Figure 6 Localization of Merlin and representative polarity complex and cell–cell junction associated proteins during collective migration.
(a) Expression pattern of Merlin within the moonlayer (Left panels), or at the leading edge (Right panels) at different time points after the confinement removal. Images show a gradual propagation of Merlin relocalization inside the monolayer with time. (b) Cytoplasmic localization of Merlin (Left—Fix1: sequential fixing and permeabilization) could not be observed in detergent-extracted samples (Right—Fix2: simultaneous fixing and permeabilization). (c) Merlin relocalization correlates with the velocity profile (representative) across the monolayer, from the leading edge towards the centre. (d) Expression pattern of Merlin upon cycloheximide treatment at 0 and 2 h after the confinement removal. Results supported Merlin relocalization over de novo synthesis of cytoplasmic Merlin during migration. (e) Localization of Par3 in stationary (Top) and migrating (Middle) wild-type MDCK cells and in Merlin-depleted cells (Bottom) in stationary condition. (f) Western blot showing Par3 depletion in MDCK cells with Par3 specific siRNAs. (g) Merlin and ZO-1 localization in control and Par3-depleted cells. Par3 siRNA2 showed similar effect. (h) Expression of E-cadherin and α-catenin in stationary and migrating MDCK monolayers. In (a–h) scale bars, 100 μm. (i) Localization of tight junction complex proteins Angiomotin and Patj in stationary and migrating MDCK cells. Scale bars, 25 μm. Panels (a-i) show the representative image of 3 independent experiments. Uncropped images of blots are shown in Supplementary Fig. 9.
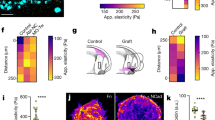
Supplementary Figure 7 The excisional 3D in vitro wound model using precast epiderm full thickness cultures and dermal equivalents.
(a) Precast epiderm full thickness (EFT) skin cultures were wounded twice with a 2 mm biopsy punch. The cultures were connected on top to a prepared dermal equivalent (DE) using 6 μl unpolymerized rat tail collagen I as ‘glue’. (b) After wounding cultures were placed into a 12-well ThinCert insert. (c) Histology of the wound cultures. H&E stained sections showed punched wounds with clear defined wound margins (arrowheads). The underlying DE was seamlessly attached providing the wound matrix. (dotted line = basement membrane). (d–e) Merlin expression in histological sections of human skin equivalent in 48 h (d) and 72 h (e) post wounding tissue. Merlin shows predominant cytoplasmic localization in migrating cells. (f) Merlin expression in histological sections of human skin shows localization to cell–cell contact. (g) MDA-MB-231 cells were used as negative control for Merlin expression. (h–j) E-cadherin localization in an unwounded (h) skin equivalent and back (i) and tongue region (j) of a wounded human skin equivalent (48 h post wounding). Scale bars, 1 cm (b), 1 mm (c), 100 μm (d-g) and 50 μm (h-j). Panels (b–j) show the representative image of 3 independent experiments.
Supplementary Figure 8 Inhibiting actomyosin based cell contractility stabilizes cell-cell junctions.
(a) FRAP curve showing E-cadherin recovery after photobleaching in E-cadherin-GFP transfected control MDCK cells (mean ± s.e.m; n = 50 cells from 3 independent experiments). (b) Comparing the FRAP curve of Blebbistatin-treated cells with that of control cells showed lower mobile fraction in former (mean ± s.e.m; n = 50 cells from 3 independent experiments in all cases). (c) Immobile fraction of E-cadherin-GFP in cells treated with DMSO (control), Blebbistatin (Bleb., 50 μM), or Y27632 (Y, 30 μM). mean ± s.e.m; n = 50 cells from 3 independent experiments in all cases (Control versus Bleb., P = 2.91 × 10−12; Control versus Y, P = 3.35 × 10−10; Wilcoxon rank-sum test). (d) Adherens and tight junction markers E-cadherin and ZO-1, respectively, in control, Blebbistatin-, or Y27632-treated cells. Scale bars, 20 μm. (e) Z-section images showing adherens and tight junctions in control or treated cells. Scale bars, 10 μm. (f) Localization of endogenous Merlin in Blebbistatin- or Y27632-treated cells. Scale bars, 100 μm.∗∗∗P < 0.001 (Wilcoxon rank-sum test). Panels (d–f) show the representative image of 3 independent experiments.
Supplementary Figure 9 Interaction with cortical actin cytoskeleton and membrane-localization stability of Merlin mutants.
(a) Schematic diagram of the FRET constructs used in Merlin-actin interaction experiments. (b) Representative image showing Merlin-cortical actin interaction in a TGFP:hMerlin-FL and TRFP:LifeAct transfected MDCK cell. Scale bars, 8 μm. (c) Mean FRET index for indicated constructs. Only E1-M was found incapable of interacting with cortical actin. (n = 7 independent experiments in all cases; hMerlin-FL versus E1-M, P = 6.67 × 10−9; Wilcoxon rank-sum test) (d) A representative FRAP curve showing the prebleach and recovery phases. Intensity was normalized to ensure average prebleach intensity = 1 and intensity immediately after bleaching = 0. Immobile fraction is the non-recovered fraction of intensity. Squares and line represent the experimental points and theoretical fitting respectively. (e) Comparison of immobile fraction for different Merlin mutants. M-EzABD showed a very high immobile fraction, while M325 was highly mobile. (n = 8 independent experiments in all cases; For hMerlin-FL versus M325, M437, hEzrin-FL, E1-M, E123-M, M-EzCC, and M-EzABD, P = 5.95 × 10−53, 8.73 × 10−38, 4.69 × 10−17, 6.53 × 10−25, 2.11 × 10−39, 4.07 × 10−36, and 1.6 × 10−63 respectively; Wilcoxon rank-sum test) Red plus sign represents the outliers that is data falling outside 2.698 times of standard deviation. ∗∗∗ P < 0.001 (Wilcoxon rank-sum test). In box plots (c,e) central mark is the median, and the edges of the box are the 1st and 3rd quartiles. Whiskers extend to the most extreme non-outlier data points. Panels b and d show the representative image of 7 and 8 independent experiments respectively.
Supplementary Figure 10 Essentiality of Rac1 activation in collective migration.
(a) Phase contrast images of wound closing by control (Left panels), Rac1 inhibitor (NSC 23766)-treated (Middle panels), Rac1-depleted (Right panels) MDCK cells at different time points after confinement removal. Both NSC 23766-treated and Rac1-depleted cells showed negligible migration activity within the investigated time frame. Scale bars, 200 μm. (b) Western blot confirming Rac1-depletion in MDCK cells with Rac1-specific siRNA. (c) Percentage of cells with cryptic lamellipodium, 2 h after removal of confinement, with first 20 layers of cells behind the leading edge. (n = 5 independent experiments; P = 4.92 × 10−8; Wilcoxon rank-sum test). In box plots central mark is the median, and the edges of the box are the 1st and 3rd quartiles. Whiskers extend to the most extreme non-outlier data points. (d) The mutant, M-EzABD, inhibits Rac1 activation even in migration promoting condition. MDCK cells, transfected as indicated, were grown to confluency for 18 h and then were allowed to migrate for 3 h. Western blots showing GTP bound Rac1 (active Rac1) and total Rac1. (e) Bars represent relative Rac1 activation results from densitometric analysis. (mean ± s.e.m; n = 3 independent experiments; P = 2.61 × 10−10; Wilcoxon rank-sum test). Statistics source data are given in Supplementary Table 3. (f) Localization of endogenous Merlin in Jasplakinolide-treated stationary and migrating cells. Even though enhanced actin binding with the M-EzABD construct prevents relocalization, stabilized cortical actin cytoskeleton does not prevent Merlin relocalization. Scale bar, 100 μm. ∗∗∗P < 0.001 (Wilcoxon rank-sum test). Panels (a, b, d, f) show the representative image of 3 independent experiments. Uncropped images of blots are shown in Supplementary Fig. 9.
Supplementary Figure 11 Polarization of lamellipodium formation and Rac1 activation in migrating epithelium.
(a–b) Representative images showing the orientations of cryptic lamellipodia in control and Merlin-depleted MDCK cells. (a) In stationary condition, Merlin-depleted MDCK cells showed higher tendency for lamellipodial protrusion formation as compared to control cells. White arrowheads indicate the lamellipodia. (b) During migration, Merlin-depleted MDCK cells showed a significantly decreased alignment in their lamellipodia as compared to control MDCK cells. White arrows approximately indicate the direction of lamellipodia with respect to the cell centre. Scale bars, 25 μm. (c) Orientations of lamellipodia with respect to the global migration direction in MDCK cells, transfected as indicated. hMerlin-FL, E1-M, and E123-M were found capable of polarizing lamellipodium formation in migrating monolayer of MDCK cells. n = 154 (Scramble control), 178 (siRNA3), 112 (hMerlin-FL), 119 (M325), 116 (M487), 108 (E1-M), 105 (E123-M), and 114 (M-EzCC) cells from 3 independent experiments. (d) Orientations of lamellipodia in control and blebbistatin-treated MDCK cells. n = 121 (control) and 117 (blebbistatin) cells from 3 independent experiments. (e) Distribution of θ (angle between Rac1 activation direction and group migration direction) in control and blebbistatin-treated MDCK cells. n = 205 (control) and 198 (blebbistatin) cells from 3 independent experiments. Panels (a, b) show the representative image of 3 independent experiments.
Supplementary Figure 12 Uncropped Western Blot images.
The corresponding main or supplementary figure numbers are shown. Blue boxes highlight the cropped segment presented in main or supplementary figures. Loading controls are provided.
Supplementary information
Supplementary Information
Supplementary Information (PDF 2262 kb)
Rights and permissions
About this article
Cite this article
Das, T., Safferling, K., Rausch, S. et al. A molecular mechanotransduction pathway regulates collective migration of epithelial cells. Nat Cell Biol 17, 276–287 (2015). https://doi.org/10.1038/ncb3115
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb3115
This article is cited by
-
Cellular mechanisms of heterogeneity in NF2-mutant schwannoma
Nature Communications (2023)
-
Reversible photoregulation of cell-cell adhesions with opto-E-cadherin
Nature Communications (2023)
-
Two Rac1 pools integrate the direction and coordination of collective cell migration
Nature Communications (2022)
-
Rac1 deficiency impairs postnatal development of the renal papilla
Scientific Reports (2022)
-
Quantifying force transmission through fibroblasts: changes of traction forces under external shearing
European Biophysics Journal (2022)