Abstract
A principal task in dissecting the genetics of complex traits is to identify causal genes for disease phenotypes. We previously developed a method to infer causal relationships among genes through the integration of DNA variation, gene transcription and phenotypic information. Here we have validated our method through the characterization of transgenic and knockout mouse models of genes predicted to be causal for abdominal obesity. Perturbation of eight out of the nine genes, with Gas7, Me1 and Gpx3 being newly confirmed, resulted in significant changes in obesity-related traits. Liver expression signatures revealed alterations in common metabolic pathways and networks contributing to abdominal obesity and overlapped with a macrophage-enriched metabolic network module that is highly associated with metabolic traits in mice and humans. Integration of gene expression in the design and analysis of traditional F2 intercross studies allows high-confidence prediction of causal genes and identification of pathways and networks involved.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
Change history
22 March 2009
NOTE: In the version of this article initially published online, there was an error in one of the genotypes in Table 5, a missing genotype in the third-to-last paragraph on page 7, and two minor typographical errors in the right column on page 6. These errors have been corrected for the print, PDF and HTML versions of this article.
References
Doss, S., Schadt, E.E., Drake, T.A. & Lusis, A.J. Cis-acting expression quantitative trait loci in mice. Genome Res. 15, 681–691 (2005).
Monks, S.A. et al. Genetic inheritance of gene expression in human cell lines. Am. J. Hum. Genet. 75, 1094–1105 (2004).
Schadt, E.E. et al. Genetics of gene expression surveyed in maize, mouse and man. Nature 422, 297–302 (2003).
Schadt, E.E. et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat. Genet. 37, 710–717 (2005).
Sieberts, S.K. & Schadt, E.E. Moving toward a system genetics view of disease. Mamm. Genome 18, 389–401 (2007).
Merkel, M. et al. Inactive lipoprotein lipase (LPL) alone increases selective cholesterol ester uptake in vivo, whereas in the presence of active LPL it also increases triglyceride hydrolysis and whole particle lipoprotein uptake. J. Biol. Chem. 277, 7405–7411 (2002).
Mirochnitchenko, O., Palnitkar, U., Philbert, M. & Inouye, M. Thermosensitive phenotype of transgenic mice overproducing human glutathione peroxidases. Proc. Natl. Acad. Sci. USA 92, 8120–8124 (1995).
Huq, A.H., Lovell, R.S., Ou, C.N., Beaudet, A.L. & Craigen, W.J. X-linked glycerol kinase deficiency in the mouse leads to growth retardation, altered fat metabolism, autonomous glucocorticoid secretion and neonatal death. Hum. Mol. Genet. 6, 1803–1809 (1997).
Chen, Y. et al. Variations in DNA elucidate molecular networks that cause disease. Nature 452, 429–435 (2008).
Ashburner, M. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 (2000).
Mi, H. et al. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 33, D284–D288 (2005).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Ghazalpour, A. et al. Genomic analysis of metabolic pathway gene expression in mice. Genome Biol. 6, R59 (2005).
Emilsson, V. et al. Genetics of gene expression and its effect on disease. Nature 452, 423–428 (2008).
Masuzaki, H. et al. A transgenic model of visceral obesity and the metabolic syndrome. Science 294, 2166–2170 (2001).
Liu, M. et al. Network-based analysis of affected biological processes in type 2 diabetes models. PLoS Genet. 3, e96 (2007).
Zhu, J. et al. Increasing the power to detect causal associations by combining genotypic and expression data in segregating populations. PLoS Comput. Biol. 3, e69 (2007).
Schadt, E.E. et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 6, e107 (2008).
Zhu, J. et al. Integrating large-scale functional genomic data to dissect the complexity of yeast regulatory networks. Nat. Genet. 40, 854–861 (2008).
Yabe, D., Brown, M.S. & Goldstein, J.L. Insig-2, a second endoplasmic reticulum protein that binds SCAP and blocks export of sterol regulatory element-binding proteins. Proc. Natl. Acad. Sci. USA 99, 12753–12758 (2002).
Janowski, B.A. The hypocholesterolemic agent LY295427 up-regulates INSIG-1, identifying the INSIG-1 protein as a mediator of cholesterol homeostasis through SREBP. Proc. Natl. Acad. Sci. USA 99, 12675–12680 (2002).
Chambers, J.C. et al. Common genetic variation near MC4R is associated with waist circumference and insulin resistance. Nat. Genet. 40, 716–718 (2008).
Frayling, T.M. et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 316, 889–894 (2007).
Loos, R.J. et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat. Genet. 40, 768–775 (2008).
Mutch, D.M. & Clement, K. Unraveling the genetics of human obesity. PLoS Genet. 2, e188 (2006).
Thorleifsson, G. et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat. Genet. 41, 18–24 (2009).
Willer, C.J. et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat. Genet. 41, 25–34 (2008).
Herbert, A. et al. A common genetic variant is associated with adult and childhood obesity. Science 312, 279–283 (2006).
Wallace, C. et al. Genome-wide association study identifies genes for biomarkers of cardiovascular disease: serum urate and dyslipidemia. Am. J. Hum. Genet. 82, 139–149 (2008).
Willer, C.J. et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 40, 161–169 (2008).
Saxena, R. et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 316, 1331–1336 (2007).
Dhanasekaran, S.M. et al. Molecular profiling of human prostate tissues: insights into gene expression patterns of prostate development during puberty. FASEB J. 19, 243–245 (2005).
Francis, K. et al. Complement C3a receptors in the pituitary gland: a novel pathway by which an innate immune molecule releases hormones involved in the control of inflammation. FASEB J. 17, 2266–2268 (2003).
Ju, Y.T. et al. gas7: A gene expressed preferentially in growth-arrested fibroblasts and terminally differentiated Purkinje neurons affects neurite formation. Proc. Natl. Acad. Sci. USA 95, 11423–11428 (1998).
Moorthy, P.P., Kumar, A.A. & Devaraj, H. Expression of the Gas7 gene and Oct4 in embryonic stem cells of mice. Stem Cells Dev. 14, 664–670 (2005).
Ishibashi, N. & Mirochnitchenko, O. Chemokine expression in transgenic mice overproducing human glutathione peroxidases. Methods Enzymol. 353, 460–476 (2002).
McClung, J.P. et al. Development of insulin resistance and obesity in mice overexpressing cellular glutathione peroxidase. Proc. Natl. Acad. Sci. USA 101, 8852–8857 (2004).
Lee, Y.S. et al. Dysregulation of adipose GPx3 in obesity contributes to local and systemic oxidative stress. Mol. Endocrinol. 22, 2176–2189 (2008).
MacDonald, M.J. Feasibility of a mitochondrial pyruvate malate shuttle in pancreatic islets. Further implication of cytosolic NADPH in insulin secretion. J. Biol. Chem. 270, 20051–20058 (1995).
van Schothorst, E.M. et al. Adipose gene expression response of lean and obese mice to short-term dietary restriction. Obesity (Silver Spring) 14, 974–979 (2006).
Qian, S. et al. Deficiency in cytosolic malic enzyme does not increase acetaminophen-induced hepato-toxicity. Basic Clin. Pharmacol. Toxicol. 103, 36–42 (2008).
Vidal, O. et al. Malic enzyme 1 genotype is associated with backfat thickness and meat quality traits in pigs. Anim. Genet. 37, 28–32 (2006).
Rahib, L., MacLennan, N.K., Horvath, S., Liao, J.C. & Dipple, K.M. Glycerol kinase deficiency alters expression of genes involved in lipid metabolism, carbohydrate metabolism, and insulin signaling. Eur. J. Hum. Genet. 15, 646–657 (2007).
Hibuse, T. et al. Aquaporin 7 deficiency is associated with development of obesity through activation of adipose glycerol kinase. Proc. Natl. Acad. Sci. USA 102, 10993–10998 (2005).
Kawai, J. et al. Functional annotation of a full-length mouse cDNA collection. Nature 409, 685–690 (2001).
Weng, L. et al. Rosetta error model for gene expression analysis. Bioinformatics 22, 1111–1121 (2006).
He, Y.D. et al. Microarray standard data set and figures of merit for comparing data processing methods and experiment designs. Bioinformatics 19, 956–965 (2003).
Storey, J.D. & Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100, 9440–9445 (2003).
Yang, X. et al. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 16, 995–1004 (2006).
Acknowledgements
The authors thank R. Davis, P. Wen, M. Rosales, X. Wu, K. Ranola and X. Xia for helping with the tissue collection. We would also like to thank L. Ingram-Drake and S. Charugundla for technical support and O. Mirochnitchenko and I. Goldberg for providing mouse models. The study was funded by US National Institutes of Health grants DK072206, HL28481 and HL30568.
Author information
Authors and Affiliations
Contributions
T.A.D., A.J.L., E.E.S., P.Y.L., X.Y., K.M.D., M.L.R. and D.J.M. designed the study. X.Y., H.Q., G.T., S. Majid, B.F., S.Q., L.W.C., D.E.-S., S. Mumick, S.S.W., A.v.N., A.G., M.M. and C.R.F. performed the experiments. X.Y., J.L.D., J. Zhu, G.T., J.K., K.W, J.R.L., T.X., M.C., J. Zhong, C.Z. and B.Z. participated in data analysis. X.Y., J.L.D., J. Zhu, S.Q. and J. Zhong wrote the manuscript, with advice and editing from T.A.D., A.J.L. and E.E.S. R.R.K. coordinated the study. T.A.D., the corresponding author, certifies that all authors have agreed to all content in the manuscript, including the data as presented.
Corresponding author
Ethics declarations
Competing interests
X.Y., J. Zhu, S.Q., J. Zhong, R.R.K., S. Mumick, T.X., M.C., C.Z., B.Z., D.J.M., M.L.R., J.L.L., P.Y.L. and E.E.S. are all employees of Merck & Co. Inc.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4, Supplementary Tables 1–14 and Supplementary Methods (PDF 703 kb)
Supplementary Figure 5
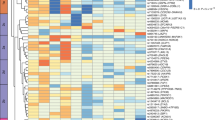
(Separate scaleable vector graphic file; open with web browser enabled with svg viewer.) The core subnetwork derived from the liver transcriptional subnetworks representative of gene expression signatures of the mouse models of the candidate genes. The liver transcriptional network is the union of Bayesian networks constructed from three crosses derived from B6, C3H and CAST. This core subnetwork consists of key regulators for fatty acid and lipid metabolism, including Insig1 and Insig2 (in red), and is enriched for genes involved in related GO biological processes. (ZIP 172 kb)
Rights and permissions
About this article
Cite this article
Yang, X., Deignan, J., Qi, H. et al. Validation of candidate causal genes for obesity that affect shared metabolic pathways and networks. Nat Genet 41, 415–423 (2009). https://doi.org/10.1038/ng.325
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.325
This article is cited by
-
NREM delta power and AD-relevant tauopathy are associated with shared cortical gene networks
Scientific Reports (2021)
-
Modulation of chromatin remodeling proteins SMYD1 and SMARCD1 promotes contractile function of human pluripotent stem cell-derived ventricular cardiomyocyte in 3D-engineered cardiac tissues
Scientific Reports (2019)
-
GWAS and enrichment analyses of non-alcoholic fatty liver disease identify new trait-associated genes and pathways across eMERGE Network
BMC Medicine (2019)
-
Integration of human adipocyte chromosomal interactions with adipose gene expression prioritizes obesity-related genes from GWAS
Nature Communications (2018)
-
Temporal genetic association and temporal genetic causality methods for dissecting complex networks
Nature Communications (2018)