Abstract
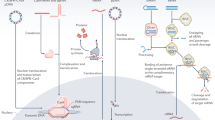
Individual genes can be targeted with siRNAs. The use of nucleic acid nanoparticles (NPs) is a convenient method for delivering combinations of specific siRNAs in an organized and programmable manner. We present three assembly protocols to produce two different types of RNA self-assembling functional NPs using processes that are fully automatable. These NPs are engineered based on two complementary nanoscaffold designs (nanoring and nanocube), which serve as carriers of multiple siRNAs. The NPs are functionalized by the extension of up to six scaffold strands with siRNA duplexes. The assembly protocols yield functionalized RNA NPs, and we show that they interact in vitro with human recombinant Dicer to produce siRNAs. Our design strategies allow for fast, economical and easily controlled production of endotoxin-free therapeutic RNA NPs that are suitable for preclinical development.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Davis, M.E. et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature 464, 1067–1070 (2010).
Tarapore, P., Shu, Y., Guo, P. & Ho, S.M. Application of phi29 motor pRNA for targeted therapeutic delivery of siRNA silencing metallothionein-IIA and survivin in ovarian cancers. Mol. Ther. 19, 386–394 (2011).
Shukla, G.C. et al. A boost for the emerging field of RNA nanotechnology. ACS Nano 5, 3405–3418 (2011).
Guo, P. The emerging field of RNA nanotechnology. Nat. Nanotechnol. 5, 833–842 (2010).
Shu, D., Shu, Y., Haque, F., Adbelmawla, S. & Guo, P. Thermodynamically stable RNA three-way junction for constructing multi-functional nanoparticles for delivery of therapeutics. Nat. Nanotechnol. 6, 658–667 (2011).
Jackson, A.L. & Linsley, P.S. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat. Rev. Drug. Discov. 9, 57–67 (2010).
Nakashima, Y., Abe, H., Abe, N., Aikawa, K. & Ito, Y. Branched RNA nanostructures for RNA interference. Chem. Commun. (Camb.) 47, 8367–8369 (2011).
Grabow, W.W. et al. Self-assembling RNA nanorings based on RNAI/II inverse kissing complexes. Nano Lett. 11, 878–887 (2011).
Khaled, A., Guo, S., Li, F. & Guo, P. Controllable self-assembly of nanoparticles for specific delivery of multiple therapeutic molecules to cancer cells using RNA nanotechnology. Nano Lett. 5, 1797–1808 (2005).
Afonin, K.A. et al. In vitro assembly of cubic RNA-based scaffolds designed in silico. Nat. Nanotechnol. 5, 676–682 (2010).
Davis, M.E. The first targeted delivery of siRNA in humans via a self-assembling, cyclodextrin polymer-based nanoparticle: from concept to clinic. Mol. Pharm. 6, 659–668 (2009).
Giljohann, D.A., Seferos, D.S., Prigodich, A.E., Patel, P.C. & Mirkin, C.A. Gene regulation with polyvalent siRNA-nanoparticle conjugates. J. Am. Chem. Soc. 131, 2072–2073 (2009).
Oh, Y.K. & Park, T.G. siRNA delivery systems for cancer treatment. Adv. Drug Deliv. Rev. 61, 850–862 (2009).
Reischl, D. & Zimmer, A. Drug delivery of siRNA therapeutics: potentials and limits of nanosystems. Nanomedicine 5, 8–20 (2009).
Kedmi, R., Ben-Arie, N. & Peer, D. The systemic toxicity of positively charged lipid nanoparticles and the role of Toll-like receptor 4 in immune activation. Biomaterials 31, 6867–6875 (2010).
Pecot, C.V., Calin, G.A., Coleman, R.L., Lopez-Berestein, G. & Sood, A.K. RNA interference in the clinic: challenges and future directions. Nat. Rev. Cancer 11, 59–67 (2011).
Tao, W. et al. Mechanistically probing lipid-siRNA nanoparticle-associated toxicities identifies Jak inhibitors effective in mitigating multifaceted toxic responses. Mol. Ther. 19, 567–575 (2011).
Shu, Y., Cinier, M., Shu, D. & Guo, P. Assembly of multifunctional phi29 pRNA nanoparticles for specific delivery of siRNA and other therapeutics to targeted cells. Methods 54, 204–214 (2011).
Jaeger, L. & Chworos, A. The architectonics of programmable RNA and DNA nanostructures. Curr. Opin. Struct. Biol. 16, 531–543 (2006).
Jaeger, L., Westhof, E. & Leontis, N.B. TectoRNA: modular assembly units for the construction of RNA nano-objects. Nucleic Acids Res. 29, 455–463 (2001).
Severcan, I. et al. Computational and experimental RNA nanoparticle design. In Automation in Genomics and Proteomics: An Engineering Case-Based Approach (eds. Alterovitz, G., Ramoni, M. & Benson, R.) 193–220 (Wiley Publishing, 2009).
Shapiro, B.A., Bindewald, E., Kasprzak, W. & Yingling, Y. Protocols for the in silico design of RNA nanostructures. In Nanostructure Design: Methods and Protocols (eds. Gazit, E. & Nussinov, R.) 93–115 (Humana Press, 2008).
Bindewald, E., Grunewald, C., Boyle, B., O'Connor, M. & Shapiro, B.A. Computational strategies for the automated design of RNA nanoscale structures from building blocks using NanoTiler. J. Mol. Graph Model 27, 299–308 (2008).
Bindewald, E., Hayes, R., Yingling, Y.G., Kasprzak, W. & Shapiro, B.A. RNAJunction: a database of RNA junctions and kissing loops for three-dimensional structural analysis and nanodesign. Nucleic Acids Res. 36, D392–D397 (2008).
Kasprzak, W., Bindewald, E., Kim, T.J., Jaeger, L. & Shapiro, B.A. Use of RNA structure flexibility data in nanostructure modeling. Methods 54, 239–235 (2011).
Martinez, H.M., Maizel, J.V. Jr. & Shapiro, B.A. RNA2D3D: a program for generating, viewing, and comparing 3-dimensional models of RNA. J. Biomol. Struct. Dyn. 25, 669–683 (2008).
Paliy, M., Melnik, R. & Shapiro, B.A. Molecular dynamics study of the RNA ring nanostructure: a phenomenon of self-stabilization. Phys. Biol. 6, 046003 (2009).
Yingling, Y.G. & Shapiro, B.A. Computational design of an RNA hexagonal nanoring and an RNA nanotube. Nano Lett. 7, 2328–2334 (2007).
Afonin, K.A., Cieply, D.J. & Leontis, N.B. Specific RNA self-assembly with minimal paranemic motifs. J. Am. Chem. Soc. 130, 93–102 (2008).
Afonin, K.A. & Leontis, N.B. Generating new specific RNA interaction interfaces using C-loops. J. Am. Chem. Soc. 128, 16131–16137 (2006).
Dibrov, S.M., McLean, J., Parsons, J. & Hermann, T. Self-assembling RNA square. Proc. Natl Acad. Sci. USA 108, 6405–6408 (2011).
Severcan, I., Geary, C., Verzemnieks, E., Chworos, A. & Jaeger, L. Square-shaped RNA particles from different RNA folds. Nano Lett. 9, 1270–1277 (2009).
Nasalean, L., Baudrey, S., Leontis, N.B. & Jaeger, L. Controlling RNA self-assembly to form filaments. Nucleic Acids Res. 34, 1381–1392 (2006).
Chworos, A. et al. Building programmable jigsaw puzzles with RNA. Science 306, 2068–2072 (2004).
Severcan, I. et al. A polyhedron made of tRNAs. Nat. Chem. 2, 772–779 (2010).
Busch, A. & Backofen, R. INFO-RNA—a server for fast inverse RNA folding satisfying sequence constraints. Nucleic Acids Res. 35, W310–W313 (2007).
Zadeh, J.N. et al. NUPACK: Analysis and design of nucleic acid systems. J. Comput. Chem. 32, 170–173 (2010).
Hofacker, I.L. et al. Fast folding and comparison of RNA secondary structures. Monatshefte f. Chemie 125, 167–188 (1994).
Elbashir, S.M., Lendeckel, W. & Tuschl, T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15, 188–200 (2001).
Hannon, G.J. RNA interference. Nature 418, 244–251 (2002).
Kim, D.H. et al. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat. Biotechnol. 23, 222–226 (2005).
Martinez, J., Patkaniowska, A., Urlaub, H., Luhrmann, R. & Tuschl, T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110, 563–574 (2002).
Rose, S.D. et al. Functional polarity is introduced by Dicer processing of short substrate RNAs. Nucleic Acids Res. 33, 4140–4156 (2005).
Bindewald, E., Afonin, K., Jaeger, L. & Shapiro, B.A. Multi-strand secondary structure prediction and nanostructure design including pseudoknots. ACS Nano published online, doi:10.1021/nn202666w (8 November 2011).
Berkhout, B. & Sanders, R.W. Molecular strategies to design an escape-proof antiviral therapy. Antiviral Res. 92, 7–14 (2011).
Liu, Y.P. et al. Combinatorial RNAi against HIV-1 using extended short hairpin RNAs. Mol. Ther. 17, 1712–1723 (2009).
Afonin, K.A., Danilov, E.O., Novikova, I.V. & Leontis, N.B. TokenRNA: a new type of sequence-specific, label-free fluorescent biosensor for folded RNA molecules. Chembiochem 9, 1902–1905 (2008).
Tyner, K. & Sadrieh, N. Considerations when submitting nanotherapeutics to FDA/CDER for regulatory review. Methods Mol. Biol. 697, 17–31 (2011).
Pulskamp, K., Diabate, S. & Krug, H.F. Carbon nanotubes show no sign of acute toxicity but induce intracellular reactive oxygen species in dependence on contaminants. Toxicol. Lett. 168, 58–74 (2007).
Vallhov, H. et al. The importance of an endotoxin-free environment during the production of nanoparticles used in medical applications. Nano Lett. 6, 1682–1686 (2006).
US Food and Drug Administration, Dept. of Health and Human Services. Investigational new drug safety reporting requirements for human drug and biological products and safety reporting requirements for bioavailability and bioequivalence studies in humans. Final rule. Fed. Regist. 75, 59935–59963 (2010).
Woods, T.O. Standards for medical devices in MRI: present and future. J. Magn. Reson. Imaging 26, 1186–1189 (2007).
Dobrovolskaia, M.A. et al. Ambiguities in applying traditional limulus amebocyte lysate tests to quantify endotoxin in nanoparticle formulations. Nanomedicine (Lond.) 5, 555–562 (2009).
Jones, C.F. & Grainger, D.W. In vitro assessments of nanomaterial toxicity. Adv. Drug Deliv. Rev. 61, 438–456 (2009).
Hall, J.B., Dobrovolskaia, M.A., Patri, A.K. & McNeil, S.E. Characterization of nanoparticles for therapeutics. Nanomedicine (Lond.) 2, 789–803 (2007).
Frantz, S. Safety concerns raised over RNA interference. Nat. Rev. Drug Discov. 5, 528–529 (2006).
Petrocca, F. & Lieberman, J. Promise and challenge of RNA interference-based therapy for cancer. J. Clin. Oncol. 29, 747–754 (2011).
Center for Biologic Evaluation and Research, Center for Devices and Radiological Health, and Center for Veterinary Medicine. Guideline on validation of the Limulus Amebocyte Lysate test as an end-product endotoxin test for human and animal parenteral drugs, biological products, and medical devices. (US Food and Drug Administration, 1987).
USP, NF 30. Bacterial Endotoxins Test. Interim revision announcement, April 1, 2011 (The United States Pharmacopeial Convention, 2011).
Bertrand, J.R., Maksimenko, A. & Malvy, C. Short double-stranded ribonucleic acid as inhibitor of gene expression by the interference mechanism. Methods Mol. Biol. 288, 411–430 (2005).
Rapozzi, V. & Xodo, L.E. Efficient silencing of bcr/abl oncogene by single- and double-stranded siRNAs targeted against b2a2 transcripts. Biochemistry 43, 16134–16141 (2004).
Li, Z.S. et al. Studies on aminoisonucleoside modified siRNAs: stability and silencing activity. Bioconjug. Chem. 18, 1017–1024 (2007).
Provost, P. et al. Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J. 21, 5864–5874 (2002).
Myers, J.W., Jones, J.T., Meyer, T. & Ferrell, J.E. Jr. Recombinant Dicer efficiently converts large dsRNAs into siRNAs suitable for gene silencing. Nat. Biotechnol. 21, 324–328 (2003).
Neun, B.W. & Dobrovolskaia, M.A. Detection and quantitative evaluation of endotoxin contamination in nanoparticle formulations by LAL-based assays. Methods Mol. Biol. 697, 121–130 (2011).
Acknowledgements
This research was supported (in part) by the Intramural Research Program of the US National Institutes of Health (NIH), National Cancer Institute, Center for Cancer Research. This work has been funded in whole or in part with federal funds from the National Cancer Institute, NIH, under contract no. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US government. This research was also supported by NIH grant no. R01GM-079604 (to L.J.). We are grateful to B. Neun for technical assistance with the LAL assay and to J. Hall for help with manuscript preparation.
Author information
Authors and Affiliations
Contributions
K.A.A., W.W.G., B.A.S. and L.J. conceived and designed the experiments. K.A.A., W.W.G., E.B., B.A.S. and L.J. contributed to the sequence and 3D model design. K.A.A., W.W.G. and F.M.W. performed self-assembly PAGE and dicing experiments. K.A.A. and F.M.W. performed DLS experiments. K.A.A., W.W.G., B.A.S. and L.J. analyzed the data. M.A.D. and K.A.A performed the LAL assay. K.A.A., W.W.G., B.A.S. and L.J. co-wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Data 1
RNA sequences used in this project. (DOCX 26 kb)
Supplementary Fig. 1
Schematic demonstration of 3′ ring scaffold functionalization with siRNA duplex. Please note that depending on tasks siRNA sense and antisense strands can be swapped. (EPS 509 kb)
Supplementary Fig. 2
Native PAGE results for native PAGE of assembly experiments for different protocols of duplex formation (between concatenated with sense strand cube strand D (siD) and corresponding radiolabeled antisense (Ant*)). (EPS 941 kb)
Supplementary Fig. 3
Native PAGE results for one-pot assembly experiments for the assembly of the cubes 3′ side concatenated with up to six identical siRNAs (eGFPS1) with corresponding yields of assembly. Radiolabeled strands are indicated with asterisks. Please note that depending on number and orientation of siRNA concatenated scaffold strands the shapes of NPs and their relative gel shifts may be slightly different (cubes functionalized with 4 siRNA duplexes). (EPS 5818 kb)
Supplementary Fig. 4
Native PAGE results for one-pot, step-wise and duplex assemblies of cubes and rings 3′ side concatenated with six different siRNAs (HIV-1). As the controls, assembled cubes and rings functionalized with 6 identical siRNAs (Figure 2) are used. (EPS 3059 kb)
Rights and permissions
About this article
Cite this article
Afonin, K., Grabow, W., Walker, F. et al. Design and self-assembly of siRNA-functionalized RNA nanoparticles for use in automated nanomedicine. Nat Protoc 6, 2022–2034 (2011). https://doi.org/10.1038/nprot.2011.418
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2011.418
This article is cited by
-
Liposome-azobenzene nanocomposite as photo-responsive drug delivery vehicle
Applied Nanoscience (2022)
-
RNA origami design tools enable cotranscriptional folding of kilobase-sized nanoscaffolds
Nature Chemistry (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.