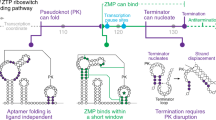
Abstract
RNAs can begin to fold immediately as they emerge from RNA polymerase. During cotranscriptional folding, interactions between nascent RNAs and ligands are able to direct the formation of alternative RNA structures, a feature exploited by noncoding RNAs called riboswitches to make gene-regulatory decisions. Despite their importance, cotranscriptional folding pathways have yet to be uncovered with sufficient resolution to reveal how cotranscriptional folding governs RNA structure and function. To access cotranscriptional folding at nucleotide resolution, we extended selective 2′-hydroxyl acylation analyzed by primer-extension sequencing (SHAPE-seq) to measure structural information of nascent RNAs during transcription. Using cotranscriptional SHAPE-seq, we determined how the cotranscriptional folding pathway of the Bacillus cereus crcB fluoride riboswitch undergoes a ligand-dependent bifurcation that delays or promotes terminator formation via a series of coordinated structural transitions. Our results directly link cotranscriptional RNA folding to a genetic decision and establish a framework for cotranscriptional analysis of RNA structure at nucleotide resolution.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Wong, T.N., Sosnick, T.R. & Pan, T. Folding of noncoding RNAs during transcription facilitated by pausing-induced nonnative structures. Proc. Natl. Acad. Sci. USA 104, 17995–18000 (2007).
Heilman-Miller, S.L. & Woodson, S.A. Effect of transcription on folding of the Tetrahymena ribozyme. RNA 9, 722–733 (2003).
Kramer, F.R. & Mills, D.R. Secondary structure formation during RNA synthesis. Nucleic Acids Res. 9, 5109–5124 (1981).
Pan, T., Artsimovitch, I., Fang, X.W., Landick, R. & Sosnick, T.R. Folding of a large ribozyme during transcription and the effect of the elongation factor NusA. Proc. Natl. Acad. Sci. USA 96, 9545–9550 (1999).
Pan, T. & Sosnick, T. RNA folding during transcription. Annu. Rev. Biophys. Biomol. Struct. 35, 161–175 (2006).
Russell, R. et al. Exploring the folding landscape of a structured RNA. Proc. Natl. Acad. Sci. USA 99, 155–160 (2002).
Al-Hashimi, H.M. & Walter, N.G. RNA dynamics: it is about time. Curr. Opin. Struct. Biol. 18, 321–329 (2008).
Garst, A.D., Edwards, A.L. & Batey, R.T. Riboswitches: structures and mechanisms. Cold Spring Harb. Perspect. Biol. 3, a003533 (2011).
Serganov, A. & Nudler, E. A decade of riboswitches. Cell 152, 17–24 (2013).
Wickiser, J.K., Winkler, W.C., Breaker, R.R. & Crothers, D.M. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol. Cell 18, 49–60 (2005).
Rieder, U., Kreutz, C. & Micura, R. Folding of a transcriptionally acting preQ1 riboswitch. Proc. Natl. Acad. Sci. USA 107, 10804–10809 (2010).
Mandal, M. & Breaker, R.R. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat. Struct. Mol. Biol. 11, 29–35 (2004).
Haller, A., Altman, R.B., Soulière, M.F., Blanchard, S.C. & Micura, R. Folding and ligand recognition of the TPP riboswitch aptamer at single-molecule resolution. Proc. Natl. Acad. Sci. USA 110, 4188–4193 (2013).
Frieda, K.L. & Block, S.M. Direct observation of cotranscriptional folding in an adenine riboswitch. Science 338, 397–400 (2012).
Wickiser, J.K. Kinetics of riboswitch regulation studied by in vitro transcription. Methods Mol. Biol. 540, 53–63 (2009).
Wong, T.N. & Pan, T. RNA folding during transcription: protocols and studies. Methods Enzymol. 468, 167–193 (2009).
Wright, D.J., King, K. & Modrich, P. The negative charge of Glu-111 is required to activate the cleavage center of EcoRI endonuclease. J. Biol. Chem. 264, 11816–11821 (1989).
Pavco, P.A. & Steege, D.A. Elongation by Escherichia coli RNA polymerase is blocked in vitro by a site-specific DNA binding protein. J. Biol. Chem. 265, 9960–9969 (1990).
Mortimer, S.A. & Weeks, K.M. Time-resolved RNA SHAPE chemistry: quantitative RNA structure analysis in one-second snapshots and at single-nucleotide resolution. Nat. Protoc. 4, 1413–1421 (2009).
Watters, K.E., Yu, A.M., Strobel, E.J., Settle, A.H. & Lucks, J.B. Characterizing RNA structures in vitro and in vivo with selective 2′-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq). Methods 103, 34–48 (2016).
Aviran, S., Lucks, J.B. & Pachter, L. RNA structure characterization from chemical mapping experiments. Proc. 49th Annual Allerton Conf. on Communication, Control, and Computing 1743–1750 (Monticello, Illinois, USA, 2011).
Bindewald, E. et al. Correlating SHAPE signatures with three-dimensional RNA structures. RNA 17, 1688–1696 (2011).
Batey, R.T., Rambo, R.P., Lucast, L., Rha, B. & Doudna, J.A. Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science 287, 1232–1239 (2000).
Komissarova, N. & Kashlev, M. Functional topography of nascent RNA in elongation intermediates of RNA polymerase. Proc. Natl. Acad. Sci. USA 95, 14699–14704 (1998).
Ehresmann, C. et al. Probing the structure of RNAs in solution. Nucleic Acids Res. 15, 9109–9128 (1987).
Baker, J.L. et al. Widespread genetic switches and toxicity resistance proteins for fluoride. Science 335, 233–235 (2012).
Ren, A., Rajashankar, K.R. & Patel, D.J. Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch. Nature 486, 85–89 (2012).
Ali, M., Lipfert, J., Seifert, S., Herschlag, D. & Doniach, S. The ligand-free state of the TPP riboswitch: a partially folded RNA structure. J. Mol. Biol. 396, 153–165 (2010).
Ottink, O.M. et al. Ligand-induced folding of the guanine-sensing riboswitch is controlled by a combined predetermined induced fit mechanism. RNA 13, 2202–2212 (2007).
Roberts, J.W., Shankar, S. & Filter, J.J. RNA polymerase elongation factors. Annu. Rev. Microbiol. 62, 211–233 (2008).
Mustoe, A.M., Brooks, C.L. & Al-Hashimi, H.M. Hierarchy of RNA functional dynamics. Annu. Rev. Biochem. 83, 441–466 (2014).
Tijerina, P., Mohr, S. & Russell, R. DMS footprinting of structured RNAs and RNA–protein complexes. Nat. Protoc. 2, 2608–2623 (2007).
Perdrizet, G.A. II, Artsimovitch, I., Furman, R., Sosnick, T.R. & Pan, T. Transcriptional pausing coordinates folding of the aptamer domain and the expression platform of a riboswitch. Proc. Natl. Acad. Sci. USA 109, 3323–3328 (2012).
Landick, R., Wang, D. & Chan, C.L. Quantitative analysis of transcriptional pausing by Escherichia coli RNA polymerase: his leader pause site as paradigm. Methods Enzymol. 274, 334–353 (1996).
Siegfried, N.A., Busan, S., Rice, G.M., Nelson, J.A. & Weeks, K.M. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP). Nat. Methods 11, 959–965 (2014).
Acknowledgements
We thank J. Roberts (Cornell University), J. Filter (Cornell University), and I. Artsimovitch (The Ohio State University) for EcoRI Gln111 protein, its expression plasmid, and thoughtful discussions. We also thank J. Liao for plasmid construction. This work was supported by the Tri-Institutional Training Program in Computational Biology and Medicine (NIH training grant T32GM083937 to A.M.Y.), a New Innovator Award through the National Institute of General Medical Sciences of the National Institutes of Health (grant no. 1DP2GM110838 to J.B.L.), the National Institute of General Medical Sciences of the National Institutes of Health (grant GM25232 to J.T.L.), a National Science Foundation Graduate Research Fellowship (grant no. DGE-1144153 to K.E.W.), and Searle Funds at The Chicago Community Trust (to J.B.L.). K.E.W. is supported as a Fleming Scholar in the Robert F. Smith School of Chemical and Biomolecular Engineering at Cornell University. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
K.E.W., E.J.S., A.M.Y., J.T.L., and J.B.L. designed experiments. K.E.W. and E.J.S. performed experiments. K.E.W., E.J.S., A.M.Y., J.T.L., and J.B.L. analyzed data. K.E.W., E.J.S., A.M.Y., J.T.L., and J.B.L. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 SHAPE-seq data for SRP RNA equilibrium refolded.
(a) SHAPE-seq reactivity matrix for all of the intermediate transcript lengths of the SRP RNA that were synthesized via E. coli RNAP and Gln111 roadblocking, extracted, denatured, and equilibrium refolded in transcription buffer before SHAPE modification. (b) Reactivity differences (∆ρ) between the cotranscriptionally and equilibrium folded SRP RNA transcript lengths. Red nucleotides are more reactive when equilibrium refolded, blue nucleotides are more reactive when cotranscriptionally folded. Due to the lack of an RNAP footprint at the 3ʹ ends of the transcripts, the equilibrium refolded lengths display transitions ~14 nt earlier than with cotranscriptional folding Results shown are n=1.
Supplementary Figure 2 Cotranscriptional DMS probing data for SRP RNA.
(a) SHAPE-seq reactivity matrix for SRP RNA (as shown in Fig. 2b, provided here for comparison with (b)). (b) SHAPE-seq reactivity matrix for all of the intermediate transcript lengths of the SRP RNA that were cotranscriptionally probed using DMS. Overall, similar transitions are observed with DMS, though the DMS data shows gaps in reactivity since it is selective for A and C bases. Reduced reactivity with DMS in the early hairpin loop (nts 9 – 19) is due to the presence of almost exclusively G and U bases in this region (Fig. 2a). Results shown are n=1 and are representative of three biological replicates (Supplementary Fig. 9b).
Supplementary Figure 3 Transcription antitermination by the B. cereus crcB fluoride riboswitch and mutants.
(a) Single-round in vitro transcription (radiolabeled) of the B. cereus crcB fluoride riboswitch. NaF was included at concentrations of 0 mM, 0.001 mM, 0.01 mM, 0.1 mM, 1 mM, and 10 mM. Terminator readthrough increases in response to fluoride. (b) The locations of mutations M18-M23 from Baker et al. (Baker, J.L. et al., Science. 335, 233-5, 2012) within the antiterminated (high fluoride) and terminated (low fluoride) secondary structures (also shown in Fig. 5a). (c) Table of mutant properties. Aptamer folding was determined from cotranscriptional SHAPE-Seq trajectories. Terminator activity was determined from part (d) and is consistent with reporter fusion assays performed in Baker et al. (Baker, J.L. et al., Science. 335, 233-5, 2012). (d) Single-round in vitro transcription (unlabeled) of the mutants shown in (b) in the presence and absence of 10 mM NaF. Transcription reactions were performed in conditions matching those used for cotranscriptional SHAPE-seq. Lane M is the RNA Century Marker (Ambion) with 100 nt and 200 nt bands shown. Templates with a functional terminator (wt, M18, M22, and M23) show a higher basal level of terminator readthrough in cotranscriptional SHAPE-Seq conditions than in conditions used for radiolabeling in (a), which contain a 10-fold lower UTP concentration. Both the wt riboswitch and the mutants follow previously described trends (Baker, J.L. et al., Science. 335, 233-5, 2012). Mutations that disrupt aptamer formation but allow terminator formation (M18 and M22) terminate at comparable efficiencies to wt without fluoride, regardless of fluoride concentration. Mutations that disrupt the terminator hairpin (M19, M20, and M21) do not terminate regardless of fluoride concentration. The combination of the M18, M19, and M20 mutations in M23 restores both aptamer and terminator hairpin base pairing and behaves as the wt riboswitch. Results shown are n=1.
Supplementary Figure 4 Folding of the P1 and P3 stems.
(a) Cotranscriptional SHAPE-seq reactivities for transcript length 40 of the B. cereus fluoride riboswitch are shown in the presence (top) and absence (bottom) of fluoride. S and L indicate P1 stem and loop nts, respectively. (b) The secondary structure of transcript length 40 is inferred from cotranscriptional SHAPE-seq reactivities, covariation analysis (Baker, J.L. et al., Science. 335, 233-5, 2012), and crystallographic data (Ren, A. et al., Nature. 486, 85-89, 2012). Nucleotide colors indicate cotranscriptional SHAPE-Seq reactivities with 10 mM NaF from (a). Gray nucleotides exist within the RNAP footprint at this length (Komissarova, N. and Kashlev, M., Proc Natl Acad Sci U S A. 95, 14699-14704, 1998). (c) Cartoon representation of P1 stem folding. Nucleotide colors correspond to key sequence elements as described in the main text and Figure 4. (d) Cotranscriptional SHAPE-Seq reactivities for transcript length 54 of the B. cereus fluoride riboswitch are shown in the presence (top) and absence (bottom) of fluoride. S and L indicate P3 stem and loop nts, respectively. (e) The secondary structure of transcript length 54 is inferred from cotranscriptional SHAPE-Seq reactivities, covariation analysis (Baker, J.L. et al., Science. 335, 233-5, 2012) and crystallographic data (Ren, A. et al., Nature. 486, 85-89, 2012). Nucleotide colors indicate cotranscriptional SHAPE-seq reactivities with 10 mM NaF from (d). Gray nucleotides exist within the RNAP footprint at this length (Komissarova, N. and Kashlev, M., Proc Natl Acad Sci U S A. 95, 14699-14704, 1998). (f) Cartoon representation of P3 stem folding is shown. Nucleotide colors correspond to key sequence elements as described in the main text and Fig. 4. Results shown are extracted from Fig. 3b.
Supplementary Figure 5 Nucleotide trace reactivity profiles for key regions of the WT fluoride riboswitch over the course of transcription.
(a) Single nucleotide trajectories displaying changes in the reactivities of nucleotides G12, G13, A14, G15, and U16 of the wt fluoride riboswitch when transcribed with either 0 mM (gray) or 10 mM NaF (black). Data for part (a), and all subsequent parts, is taken from the cotranscriptional SHAPE-Seq matrices in Fig. 3b. (b) Single nucleotide trajectories displaying changes in the reactivities of nucleotides A24, A25, and C27 of the wt fluoride riboswitch when transcribed with either 0 mM (gray) or 10 mM NaF (black). Positions U23 and A26 in this region show low reactivities that persist throughout transcription. (c) Single nucleotide trajectories displaying changes in the reactivities of nucleotides A52, G53, U54, and A55 of the wt fluoride riboswitch when transcribed with either 0 mM (gray) or 10 mM NaF (black). These nucleotides are found on the upper 5’ side of the terminator. (d) Single nucleotide trajectories displaying changes in the reactivities of nucleotides A67, G68, G69, A70, G71, and U72 of the wt fluoride riboswitch when transcribed with either 0 mM (gray) or 10 mM NaF (black). These nucleotides comprise the ribosome binding site (RBS) found in the 3’ side of the terminator stem. Results shown are extracted from Fig. 3b.
Supplementary Figure 6 Cotranscriptional SHAPE-seq data for fluoride riboswitch mutants M18 and M22 with disrupted pseudoknots.
(a) Reactivity matrices of the M18 mutant transcribed with either 10 mM (top) or 0 mM NaF (bottom). (b) Reactivity differences (∆ρ) between the matrices in (a). Positions that are red are more reactive when fluoride is present, while positions that are blue are more reactive without fluoride. (c) Single nucleotide trajectories (taken from (a)) displaying changes in the reactivities of key nucleotides, or nucleotides representing key regions, when transcribed with either 0 mM (gray) or 10 mM NaF (black). (d) As in (a), but for mutant M22. (e) Reactivity differences (∆ρ) between the matrices in (d). (f) Single nucleotide trajectories (taken from (d)) displaying changes in the reactivities of key nucleotides, or nucleotides representing key regions, when transcribed with either 0 mM (gray) or 10 mM NaF (black). Results shown are n=1.
Supplementary Figure 7 Cotranscriptional SHAPE-seq data for fluoride riboswitch mutants M20 and M21 with terminator-stem disruptions.
(a) Reactivity matrices of the M20 mutant transcribed with either 10 mM (top) or 0 mM NaF (bottom). (b) Reactivity differences (∆ρ) between the matrices in (a). Positions that are red are more reactive when fluoride is present, while positions that are blue are more reactive without fluoride. (c) Single nucleotide trajectories (taken from (a)) displaying changes in the reactivities of key nucleotides, or nucleotides representing key regions, when transcribed with either 0 mM (gray) or 10 mM NaF (black). (d) As in (a), but for mutant M21. (e) Reactivity differences (∆ρ) between the matrices in (d). (f) Single nucleotide trajectories (taken from (d)) displaying changes in the reactivities of key nucleotides, or nucleotides representing key regions, when transcribed with either 0 mM (gray) or 10 mM NaF (black). Results shown are n=1.
Supplementary Figure 8 SHAPE-seq data for WT fluoride riboswitch equilibrium refolded.
(a) SHAPE-seq reactivity matrix for all of the intermediate transcript lengths of the wt fluoride riboswitch that were first transcribed with 10 mM NaF, then extracted, denatured, and equilibrium refolded in transcription buffer containing either 10 mM (top) or 0 mM NaF (bottom) before SHAPE modification. (b) Reactivity difference (∆ρ) matrix between the matrices in (a) as described in Figure 3c. Few differences are observed between the refolding conditions, except for generally higher reactivities in the P1 loop (nts 12-16) without ligand (although both are high) and the appearance of high reactivity at position A22 across lengths 47-66. A sharp increase in reactivity is observed in the P1 loop at length 68 indicating that the terminated structure (PK1 open) is thermodynamically more stable in equilibrium, independent of the presence of fluoride. The differences observed between the cotranscriptional and equilibrium refolding matrices when fluoride is present provides strong evidence that the fluoride riboswitch can only function cotranscriptionally. Because the RNAP footprint is absent in equilibrium refolding, all of the transitions are observed ~14 nt earlier (Komissarova, N. and Kashlev, M., Proc Natl Acad Sci U S A. 95, 14699-14704, 1998). Results shown are n=1.
Supplementary Figure 9 Additional replicates of cotranscriptional SHAPE-seq data for the E. coli SRP RNA and B. cereus crcB fluoride riboswitch.
Additional two replicates each for SRP RNA cotranscriptional SHAPE-seq data using BzCN (a) or DMS (b). Additional replicates for the crcB fluoride riboswitch cotranscriptional SHAPE-seq data with (c) 10 mM NaF and (d) 0 mM NaF. Results shown are n=1.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–9 Supplementary Tables 1–4 and Supplementary Note (PDF 2430 kb)
Supplementary Table 5
Reverse oligonucleotides for IVT template PCR (XLSX 61 kb)
Source data
Rights and permissions
About this article
Cite this article
Watters, K., Strobel, E., Yu, A. et al. Cotranscriptional folding of a riboswitch at nucleotide resolution. Nat Struct Mol Biol 23, 1124–1131 (2016). https://doi.org/10.1038/nsmb.3316
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.3316
This article is cited by
-
Probing the dynamic RNA structurome and its functions
Nature Reviews Genetics (2023)
-
Structure, folding and flexibility of co-transcriptional RNA origami
Nature Nanotechnology (2023)
-
Observation of structural switch in nascent SAM-VI riboswitch during transcription at single-nucleotide and single-molecule resolution
Nature Communications (2023)
-
Programmable single-stranded architectures for computing
Natural Computing (2023)
-
Spacer prioritization in CRISPR–Cas9 immunity is enabled by the leader RNA
Nature Microbiology (2022)